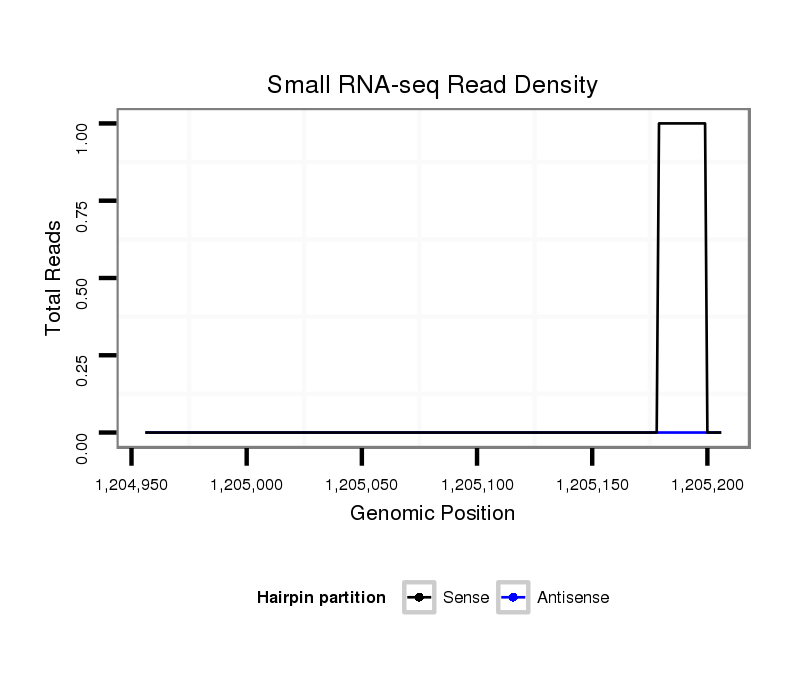
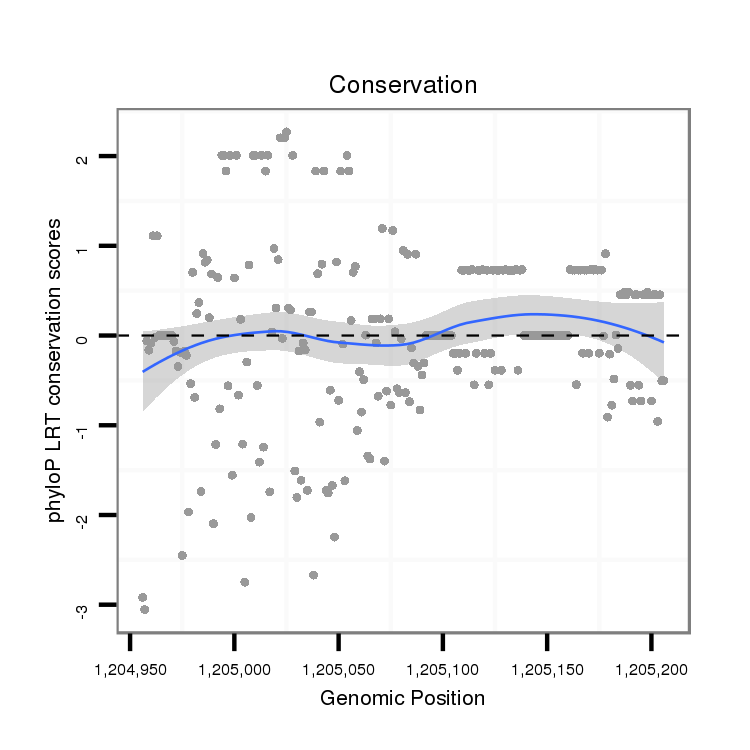
| TGTGACTCT-------GGCTGGGAAGACGGCGGAAGCGAATCTGCTAATAATGCCGACCATGTTGGACTATGTCATCCTCGGCATGGACTTTCTATGCGCCATCGGCACTACGGTGCGTTGCGGCAACACAGAGCTGGAGATGAGGATGGTGGACGACGTGGCAGAAGGAGCGTCGCTGTCGAGTCGACAAAGAGTGGAGAAAGGCATCAGTGGTTTTAGAGGGCAAGACCAGC------------------------------------------------------------------------------------------------TCGCCAAAGGAGATGCGAAGGAGTCGGGCC | Size | Hit Count | Total Norm | Total | M046
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGACG............................................................................................................................................................................................................ | 22 | 17 | 0.53 | 9 | 8 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGAC............................................................................................................................................................................................................. | 21 | 17 | 0.35 | 6 | 5 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGACGA........................................................................................................................................................................................................... | 23 | 17 | 0.35 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGA.............................................................................................................................................................................................................. | 20 | 17 | 0.35 | 6 | 4 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................TCGGCATGGACTTTCTATGCGCCATC................................................................................................................................................................................................................................................................ | 26 | 11 | 0.18 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................TGGAGATGAGGATGGTGGACGA........................................................................................................................................................................................................... | 22 | 17 | 0.18 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGG............................................................................................................................................................................................................... | 19 | 18 | 0.17 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TCGGCATGGACTTTCTATGCGCCATCGGC............................................................................................................................................................................................................................................................. | 29 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGACGAC.......................................................................................................................................................................................................... | 24 | 16 | 0.13 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................TGGACTTTCTATGCGCCATCGGCACTAC........................................................................................................................................................................................................................................................ | 28 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TGGAGATGAGGATGGTGG............................................................................................................................................................................................................... | 18 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TCGGCATGGACTTTCTATGCGCCATCGG.............................................................................................................................................................................................................................................................. | 28 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TCGGCATGGACTTTCTATGCGCCATCG............................................................................................................................................................................................................................................................... | 27 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TCGGCATGGACTTTCTATGCGCC................................................................................................................................................................................................................................................................... | 23 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TCATCCTCGGCATGGACTTTCTATG....................................................................................................................................................................................................................................................................... | 25 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TTAGAGGGCAAGACCAGC------------------------------------------------------------------------------------------------TCGC.......................... | 22 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGACGAA.......................................................................................................................................................................................................... | 24 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................CTGGAGATGAGGATGGTGGAAT............................................................................................................................................................................................................ | 22 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................TGGAGATGAGGATGGTGGAC............................................................................................................................................................................................................. | 20 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TGGAGATGAGGATGGTGGACG............................................................................................................................................................................................................ | 21 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTGGAGATGAGGATGGTG................................................................................................................................................................................................................ | 18 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TATGCGCCATCGGCACTACGGTGCGTTG............................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GAGATGAGGATGGTGGACGACGTGG...................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TGAGGATGGTGGACGACGTGG...................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |