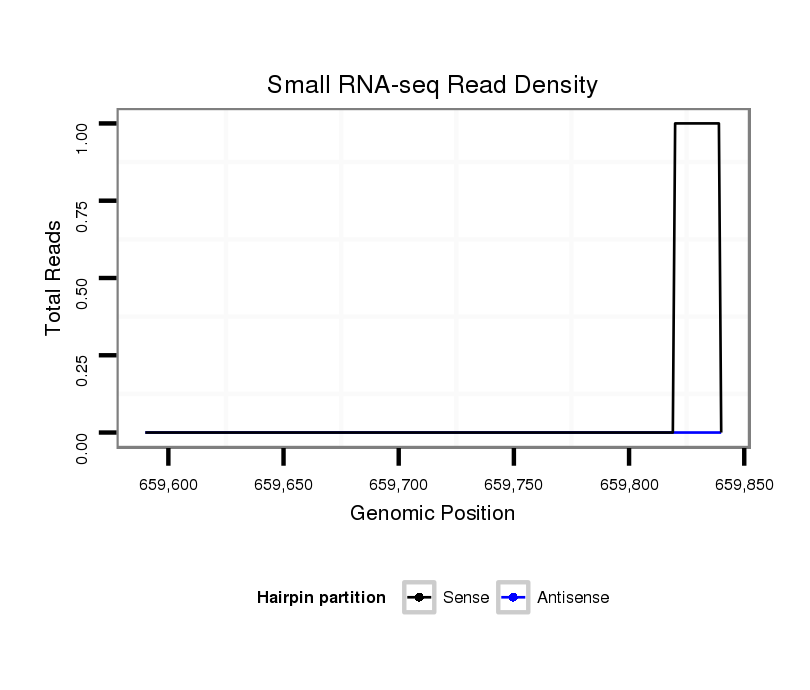
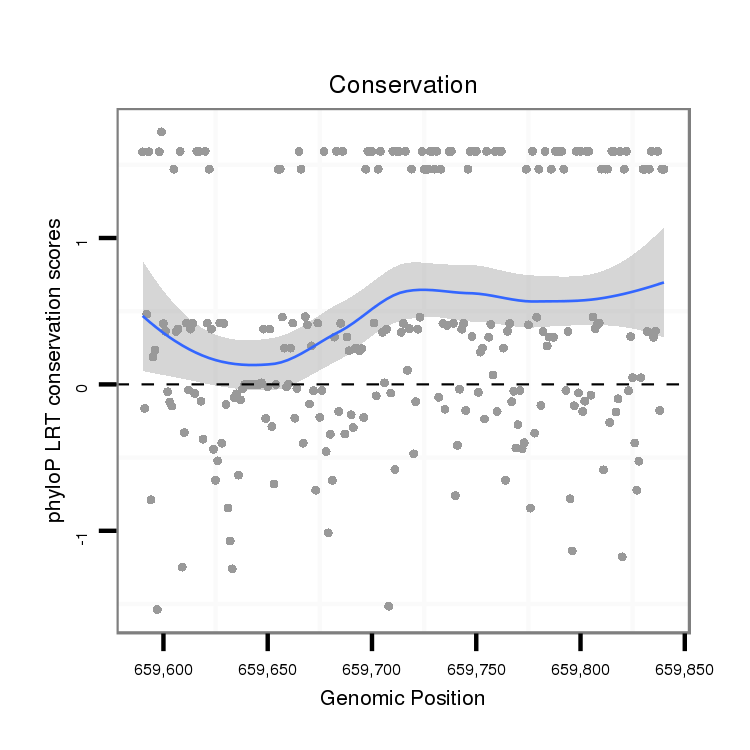
ID:dvi_9680 |
Coordinate:scaffold_12958:659640-659790 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ11164-cds]; exon [dvir_GLEANR_11128:2]; intron [Dvir\GJ11164-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GAGCCCTACAGCTATGCGTACTGGTTTTCATACGGATTCATGCGATGAAGGTAAGATTCGACGGGGGACTGGATTTGCAGAAGATACTGCCCCTTTAACATAGCAAAGAAATAGATGTGATTTTTTTTCGGATATGCCAAGAAGGAGTTATATATAGAGATTACCTTCTTTAAGTCGAGGCGCCGCTTAACCCATCGTAAAAGCCTGCTGTACAAATGCTCCCCTTATATAGATGACTCAGGCGTTCTACG **************************************************........(((.(((((.((.((((.((((......))))..((((........))))......((((((((((((((.((.....)).)))))))))))))).((((((......)))))))))).))...))......))).)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060671 9x160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060660 Argentina_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060689 160x9_testes_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TGCCAAGAAGGAGTTATATATAGAGATC......................................................................................... | 28 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................TTTTCATACGGATTCATGCGATGAAGGC....................................................................................................................................................................................................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AGATGACTCAGGCGTTCTAC. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GTTATTTTTTTTCGGATA..................................................................................................................... | 18 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................CGCGGCGCCGCTTTACCC.......................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................GCGGGACCGGATTTGCGGA.......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .AACCCTACGGATATGCGT........................................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................CACTGGATCTGCAGAAGAAA..................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................TTGTGGCGGCGCTTAACCC.......................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
CTCGGGATGTCGATACGCATGACCAAAAGTATGCCTAAGTACGCTACTTCCATTCTAAGCTGCCCCCTGACCTAAACGTCTTCTATGACGGGGAAATTGTATCGTTTCTTTATCTACACTAAAAAAAAGCCTATACGGTTCTTCCTCAATATATATCTCTAATGGAAGAAATTCAGCTCCGCGGCGAATTGGGTAGCATTTTCGGACGACATGTTTACGAGGGGAATATATCTACTGAGTCCGCAAGATGC
**************************************************........(((.(((((.((.((((.((((......))))..((((........))))......((((((((((((((.((.....)).)))))))))))))).((((((......)))))))))).))...))......))).)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................ATTTATATGTCTAATGTAAGAAA................................................................................ | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 |
| ..........................................................................................................................................................................ATTCAGCTCCGAGGCTAC............................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12958:659590-659840 + | dvi_9680 | GAGCCCTACAGCTATGCGTACTGGTTTTCATACGGATTCATGCGATGAAGGTAAGATTCGACGGGGGACTGGATTTGCAGAAGATACTGCCCCTTTAACATAGCAAAGAAATAGATGTGATTTTTTTTCGGATATGCCAAGAAGG--AGTTATATATAGAGATTACCTTCTTTAAGTCGAGGCGCCGCTTAACCCATCGTAAAAGCCTGCTGTACAAATGCTCCCCTTATATAGATGACTCAGGCGTTCTACG |
| droWil2 | scf2_1100000003160:1334-1577 - | GGGCGGTACAAAAACGAATTGAAAACTTTTTGCAAGGCATGTGTAAAAG--------GTCTTGC-GGAGAACGACTGAAGCACAATTTGGCTATAAACAGCGATGTGGAAACCGATTCAATGTTTATTCGTGCATGCCAAGAAGAGGAGTTTCCAGAAGAGATCAGATGCTTAAACCAAGGACATGGCATAACAAATAGGAAAAGTAAGCTATTTAAGTGCACTCCTTACCTGGACGAGTTAGGAGTGCTACG | |
| droAna3 | scaffold_13744:58567-58799 + | GAGCC---CAAAGGCGAGTGCTGGATTTCATACGATATATTCAAGTTCG--------ACAGCCT-GGACCTGAATTGAAAAGAATACTCGAACTAAAAC--------GAAATGGATACGGTCTTTATTCGAGCTTGCCAAGAGGAAACGTTTATAGATGAAATTCGCTGCTTAAAGAAAGATGGTCGTGTCTCAAACCGGAAAAGCAGCGTATTCAAATGTTCGCCGTATCTTGACGAGTCAGGAGTGATTCG | |
| droKik1 | scf7180000302237:1087-1330 + | GTGCTGCCCAAAAACGAGTACTGGATTTCCTACGCTATATATGCAAGGA--------ACCGCGT-GGGCCTGAACTGGTGAGACTATTCAAACTAAAACGCTATGTGGAAATGGATACTGTCTTTATACGAGCATGCCAAGAAGAAGCGTTTGCTGATGAAATACGCTGTCTAAAGAAAGGAGACTGCGTAACTAACCGGAAAAGTACCATTTTCAAGCTCTCACCATATCTTGATGCAGCCGGAGTGCTTCG | |
| droRho1 | scf7180000772251:375-618 + | GTGCTGCGCAAAAGAGAGTTTTGGATTTTCTACGCCTTCTTTCCGTGAA--------ACCGCAC-GGACCTGAGCTGGAGGGACTGCTTTTGCTTAAGCGCGATGTAGAAATGGACACAATTTTCATACGGACATGCCAAGAAGAAGAGTTTTTTGATGAAATTCGATGCCTTAAGGAAAATCGTTGCCTGACAAACCGGAAAAGCATAATATATAAATGCTCGCCGTACTTAGACCCAGTCGGAGTGCTTCG | |
| droTak1 | scf7180000413896:612-855 + | GTGCTGCCCAAAAAAGAGTGTTAGACTTTCTACGCCTTCTTCCTGTAAA--------ACCGCAT-GGACCTGAGCTGGAGAGATTGCTTTTGCTTAAGCGTGATGTGGAAATGGACACCATTTTCATAAGGACATGCCAAGAGGAAGAATTTTTTGACGAAATTCGATGCCTTAAGGAAAATCGCTGCCTGACAAACCGGAAAAGCATTATATATAAATGCTCGCCGTACCTGGACCCAAACGGAGTGCTCCG | |
| droEre2 | scaffold_4929:25547053-25547062 - | GTTCCGCACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:46 PM