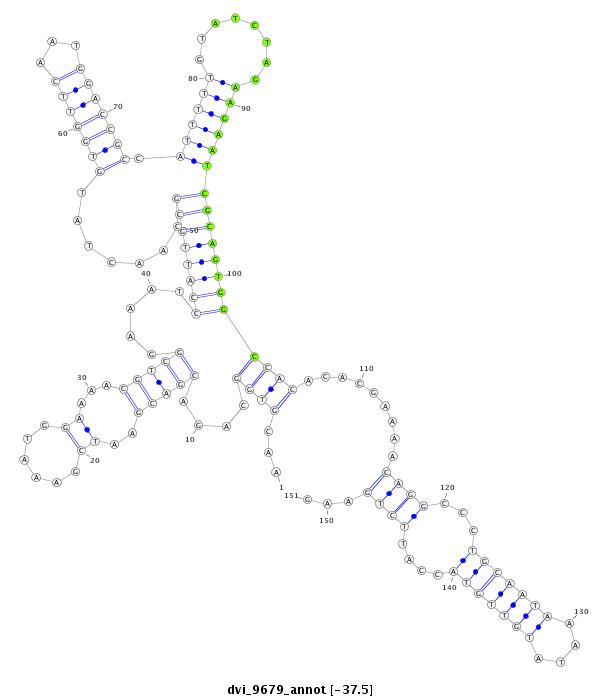
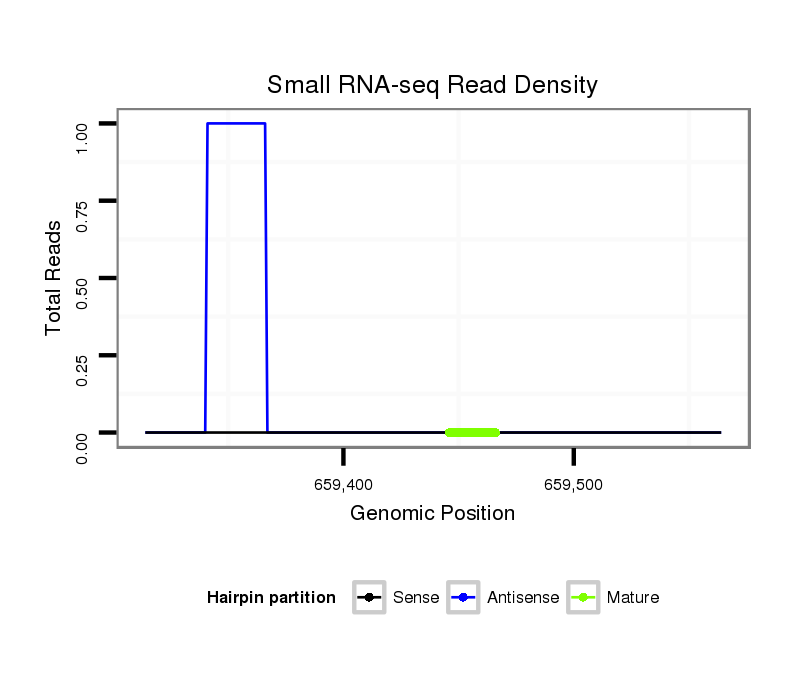
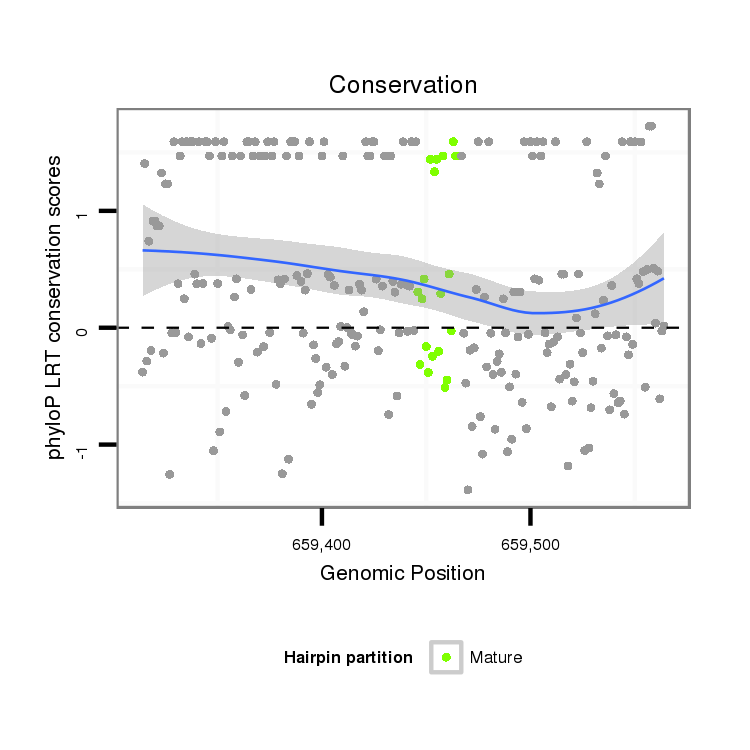
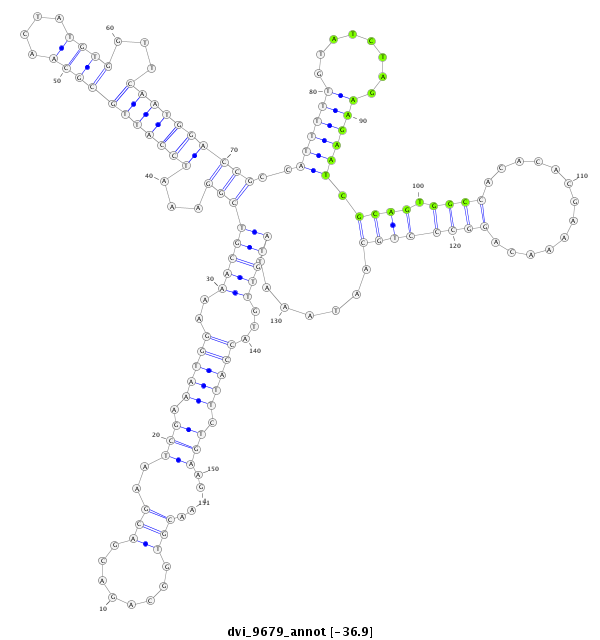
ID:dvi_9679 |
Coordinate:scaffold_12958:659364-659514 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.3 | -36.9 | -36.9 |
 |
 |
 |
exon [dvir_GLEANR_11128:2]; CDS [Dvir\GJ11164-cds]; intron [Dvir\GJ11164-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGATATAGCTTCGCATCAGACAGATTTTAGAAGAATCAGATCAGAATCAAAACGTGGCAGACGACGAATCGAAATGGAAAACGTCGGAAATCCATTGCGCAACTATGTGGTTCAATGGACCGCCATTTTTGTATCTAGAAGAATCGCAGTGGCCACACACGAAAACAGGCCCTGCAATAAATATGTTGTACCATTCTGAAGAACAATTCAATCACACGTCGTCCTGGAGATGTTTCATGCCGGATCTGCAG **************************************************...((((....(((((..((......))...))))).....((((((((.......(((((((...))))))).((((((........)))))))))))))))))).........((((...(((((((....)))))))....))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060685 9xArg_0-2h_embryos_total |
SRR060656 9x160_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............CATCAGACAGACTTTGGAAGA......................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGACGACGAATCGAAAT................................................................................................................................................................................ | 18 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................ATCTAGAAGAATCGCAGTGGC.................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................GTCCTGGAGTTGATTGATG............ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................ATCACACGTCGTCCTGGAGAT.................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................TGTATCTAGAAGAATCGCAGT..................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AATCACACGTCGTCCTGGAGA..................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ACTATATCGAAGCGTAGTCTGTCTAAAATCTTCTTAGTCTAGTCTTAGTTTTGCACCGTCTGCTGCTTAGCTTTACCTTTTGCAGCCTTTAGGTAACGCGTTGATACACCAAGTTACCTGGCGGTAAAAACATAGATCTTCTTAGCGTCACCGGTGTGTGCTTTTGTCCGGGACGTTATTTATACAACATGGTAAGACTTCTTGTTAAGTTAGTGTGCAGCAGGACCTCTACAAAGTACGGCCTAGACGTC
**************************************************...((((....(((((..((......))...))))).....((((((((.......(((((((...))))))).((((((........)))))))))))))))))).........((((...(((((((....)))))))....))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060663 160_0-2h_embryos_total |
GSM1528803 follicle cells |
M027 male body |
SRR060668 160x9_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR1106722 embryo_10-12h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................ATCTTCTTAGTCTAGTCTTAGTTTTG...................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GCTAAGTTAGTGTACAGCA............................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TACCCTTTGCAGACTTTAG............................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CGACGAAGTAAGGCCTAGAC... | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TTGCATAGTCTGCTGCTCA...................................................................................................................................................................................... | 19 | 3 | 15 | 0.20 | 3 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| .......................................................................................................................CGCGATAACAACATAGATC................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATAGGTAACGCGTTG.................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................TTCTTAGTCTAGCCTTCT........................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12958:659314-659564 + | dvi_9679 | TGAT--ATAGC--TTCGCATCAGACAGATTTTAGAAGAATCAGAT------------------------CAGAATCAAAACGTGGCAGACGACGAATCGAAATGGAAAA---CGTCGGAAATCCATTGCGCAACTATGTGGTTCAATGGACCGCCATTTTTGTATCTAGAAGAATCGCAGTGGCCACAC-ACGAAAACAGGCCC---------TGC--AATAAATATGTTGTACCATTCTGAAGAACAATTCAATCACACGTCGTCCTGGAGATGTTTCATGCCGGATCTGCAG |
| droWil2 | scf2_1100000006827:373-632 - | CG---------------TGTTGGAGAAATTCTAAAAGACTTTGACGTACTATCTTGGTGAT--------ATGATGCATAACGTTGCAGACGATGGCACGAAATGGGCAAAGATGCCAGACATGACCGGGAATTCAAGGTGGTTTGTTGGCCCGAGATTCTTGTATGAACAAGAGACACAATGGCCTATCATGGAGCAGAGCCCGAGCAAGA---CGAGTGAAAGTATCTTGAACCACGAGTAGAAAGTAAA-AAGGAA-------CTCTAGTTTGCATCACACCTGATCCGGAA | |
| droAna3 | scaffold_12947:1088840-1089118 - | C------------TACGAGTCGGAGAAATCCTGGAAGGATCGGAGATTGTCGATTGGCGCTGGATACCTTCGGAGCTTAATGTGGCGGACGAAGGAACCAAATGGACAAAAAACTGTGAAATGACCAGCTCGATAAGGTGGTTTACTGGTCCCGAATTCTTATTGCAGAACGAGTCAAGATGGCCGTCA-CAAATCGGAACCGCTGTGAGCTCCGA--TGATACACTGCTGTATCACGCATATATGGTGCGGATGGACCCGTCACCTATTGCCTCTATTATGCCAAATCCGCTA | |
| droKik1 | scf7180000297786:21960-22235 + | TGG--------CTTACGTATTGGAGAGATTCTTGAAGAATCTGAGATTGAAGATTGGCGCTGGGTACCTTCGGAACAGAATGTGGCGGATGAAAGGACCAAATGGACTAAGAATTGCGAAATAAACAGCTCAGGAAGATGGTTTACTGGCCCGGAATTTTTATTGCAG-------CTCAGTGGCCATCG-CGAACAGGAGTTCACATTAGTAGCGA--GTGTGAACTAATGTACCATGATGAGGAAACGTGGGTGAACAAGTCACCTTTTGATTCGATTATGCCGGATCCGTTG | |
| droRho1 | scf7180000776979:11098-11383 - | TG-----TAGCTTTACGAGTCGGAGAAATTCTTGAAGGCTCCGAAGTTGGTGAATGGCGCTGGGTACCCTCGGAACAAAATGTAGCAGACGATGGGACTAAACGAACAAAAAGTGTTGAGAGAAATAGTTCAATTCGATGGTTTTCCGGACTGAATTATCTGTTTCAAGATGAATCGGAGTGGCCACGC-GGAAATGAAATGTCTAAAACCAGAGA--AACTCAAGTATTGTACCACAAGGAGAAAATTAAGAAACAAATCTCGCCTTTGTCATGTGTAATGCCGGATTCACTA | |
| droTak1 | scf7180000413361:7012-7302 - | CGATTCGTAGCTTTGCGGGTCGGAGAAATTCTTGAAGGTTCCGAAGTTGGTGAATGGCGCTGGGTACCCTCGGAACTAAATGTAGCGGATGATGGTACAAAATGGTCAAAAGGTGTTGAGGTAAATAGTTCAATACGATGGTTTTCCGGACCGAATTATCTGTTGCAGGATGAGTCGGAGTGGCCACTC-GGAAAAGAAATATCTAAAGCCAGAGA--GACTCAAGTGTTGTACCACAAGGAGGAAAGTAAGAAACAGGTCTCGCCGCTGGCATCTGTTATGCCGGATCCATTA | |
| droEre2 | scaffold_4929:25547088-25547101 - | TG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACGATCTACAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:51 PM