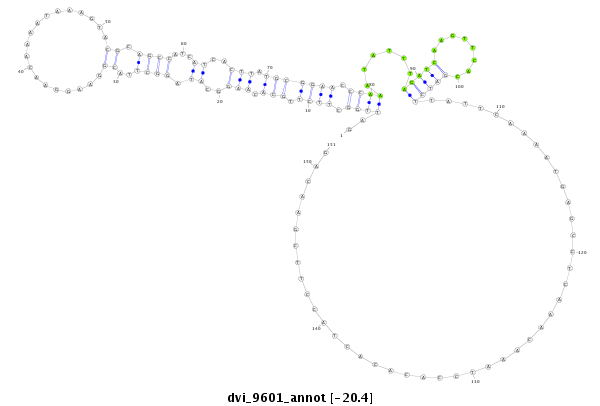
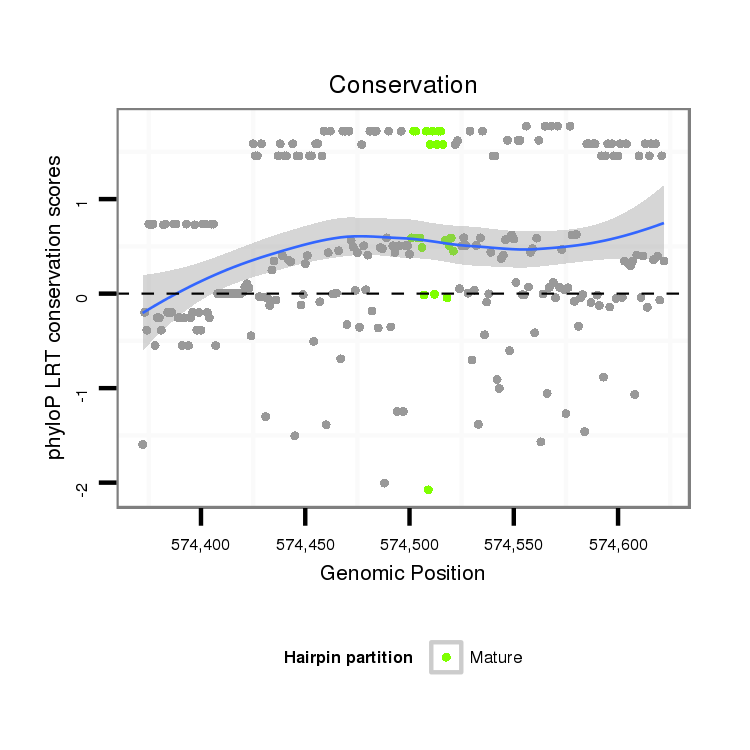
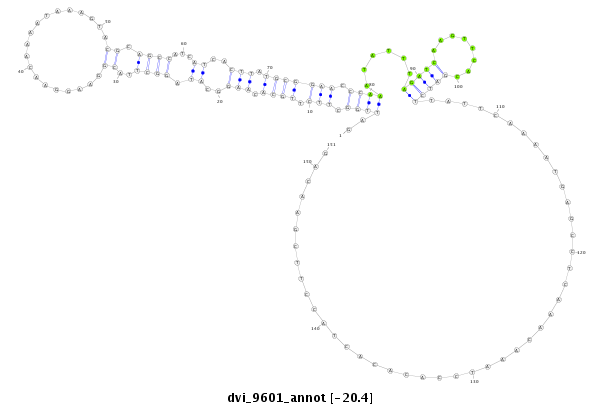
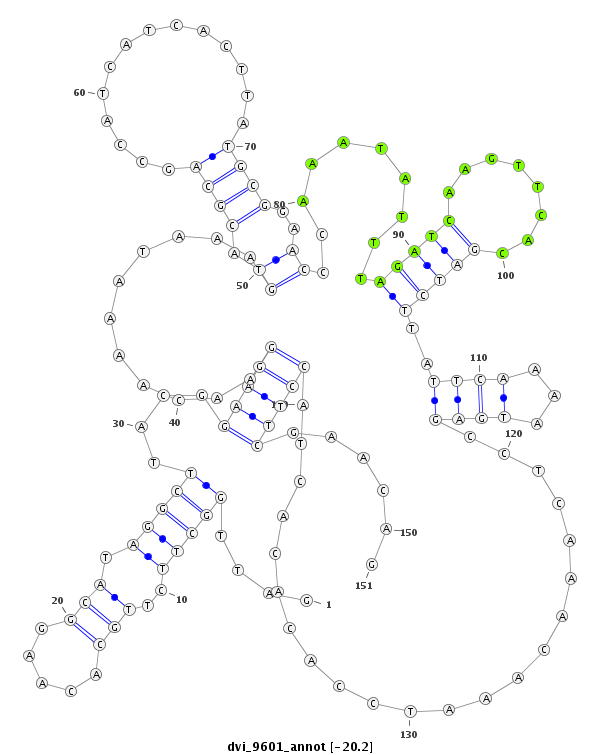
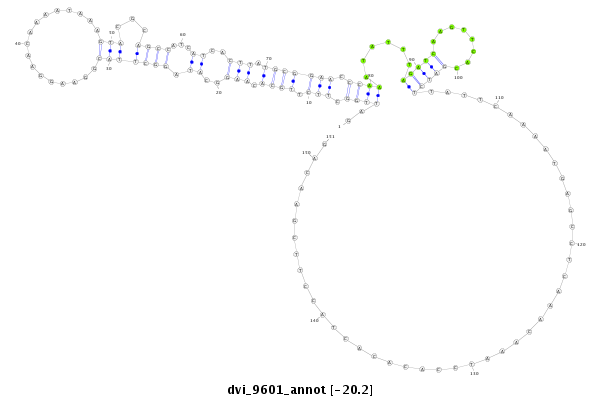
ID:dvi_9601 |
Coordinate:scaffold_12936:574422-574572 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.4 | -20.2 | -20.2 |
 |
 |
 |
exon [dvir_GLEANR_14041:3]; CDS [Dvir\GJ14136-cds]; intron [Dvir\GJ14136-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCTCAGTTCCACGAAATACATGACCCTTAAAATACGAAAAGGAGGATGAGATTGGCTTCTTGCACAAGGCATAGGCTTACGGAAGGAACAAAATAAAGTACGCAGCCATCATCACTTATGCGGAACCCAAATATTTAGATCAAGTTCACGATCTTATTCAAAATGAGCCTCAAACAAATCCACACACTACCTTCGAACAGGTCTTTCTTAAAAATTTCTCTGAGTCAGAGGTATCACGCCTCGACAAACC **************************************************..((((.((((.(((.(((..((.((((..((...................)).))))...))..))).))))))).))))......(((((........)))))..............................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................TCTTAAAAATTTCTCTGAGTC........................ | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................AAATATTTAGATCAAGTTCAC..................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................GAAACTGAGGATGAGATT..................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ....................................GAAAAGGAGGAAGAGATA..................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
ACGAGTCAAGGTGCTTTATGTACTGGGAATTTTATGCTTTTCCTCCTACTCTAACCGAAGAACGTGTTCCGTATCCGAATGCCTTCCTTGTTTTATTTCATGCGTCGGTAGTAGTGAATACGCCTTGGGTTTATAAATCTAGTTCAAGTGCTAGAATAAGTTTTACTCGGAGTTTGTTTAGGTGTGTGATGGAAGCTTGTCCAGAAAGAATTTTTAAAGAGACTCAGTCTCCATAGTGCGGAGCTGTTTGG
**************************************************..((((.((((.(((.(((..((.((((..((...................)).))))...))..))).))))))).))))......(((((........)))))..............................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060654 160x9_ovaries_total |
SRR060689 160x9_testes_total |
SRR060673 9_ovaries_total |
V047 embryo |
V116 male body |
V053 head |
SRR060686 Argx9_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................ACCGAAGAACGTGTTCCGTAT................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................TTTAGGGGTGTGATGGTAGC....................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................TTTAGGGGTGTGATGGTAG........................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GTGATTTATGGGCTGGGAAT............................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................CGGAAGTAGTGAGTGCGCC............................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GTCTTGGGTTTCTGAATCTA.............................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........AGTTGCTTTAGGTACTGGGG............................................................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AAAGAATTTTCAGTGAGACT........................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................TTAAAGCGACTCAGGGTCC................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................GGGTTCCGTAGCCGCATG.......................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........CGGCTTCATGTACTGGGA............................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TAATAGTGAAGACGCATTG............................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12936:574372-574622 + | dvi_9601 | TGCTCAGTTCCACGAAATACATGACCCTTAAAATACGAAAAGGAGGATGAGATTGGCTTCTTGCACAAGGCATAGGCTTACGGAAGGAACAAAATAAAGTACGCAGCCATCATCACTTATGCGGAACCCAAATATTTAGATCAAGTTCACGATCTTATTCAAAATGAGCCTCAAACAAATCCACACACTACCTTCGAACAGGTCTTTCTTAAAAATTTCTCTGAGTCAGAGGTATCACGCCTCGACAAACC |
| droMoj3 | scaffold_1712:2278-2480 + | G-----------------------------------------------AAGGCTGGGTTGAGCTGCAGGGCATCGGTACGCGTAAAGAGCAC-GAGAAGTACACTGCCGTTATAACCTACGCAGATCCAAAATATCTGGAGCAAGTGCATGATCTAGTAGTGAACGAGCCAACTTCAAATCCATACACCACGTTGAAACAATCAATTCTCGCCAAATTCGCAGAATCTGAAATGGTTCGTCTTGATAAGCT | |
| droPer2 | scaffold_7:4418789-4418828 + | A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAATCCAGATAAAGCTTTTGAAAAGGTCTTTCTGAAA-------------------------------------- | |
| droAna3 | scaffold_13333:210760-210997 + | CAATCACAAACATAGAAATGTTCTTCACGAAGTTAG------------AAAGTTGGTTCGCACTACAAGGACTTGGTGCACGAAAAGAACAG-GAAAAGTTCGCTGCAGTCATCGCCTACGCAGACCCCAAATACCTGGAACAAGTACACGATCTTGTAAACAATCCACCCGAAACAGCCCCTTATTCCACCTTAAAGGAAGCGATCCTTTCAAAGTTTACTGACTCGGAAATGGTACGTTTGGACAGACT | |
| droKik1 | scf7180000299059:4881-5077 + | -----------------------------------------------------TGGTTTTTGCTGCAAGGCCTTGGCGCACGCAAAGAACAA-GAGAAATATGCGGCAGTAATCGCGTACGCCGATCCCAAATACCTCGATCAAGTGCACGATCTTGTCAATAACCCACCACAAACGAATCCGTACTCCACACTTAAACAAGCAATTTTGTCTAAATTTGCCGAATCAGAGATGGTGCGCCTCGACAGGCT | |
| droRho1 | scf7180000778177:2703-2904 - | G-----------------------------------------------AGAGCTGGTTCATGCTGCAAGGCCTAGGTGCACGCAAAGAGCAA-GAAAAATACGCAGCAGTAATCGCGTACGCTGATCCCAAATACCTGGATCAAGTGCACG-CCTTGTCAACAACCCACCACAAACGAATCCGTACTCCACCTTTAAACAAGCCATTAAGTCCAAATTCGCAGAATCAGAGATGGTGTGCCTCGACAGACT | |
| droBia1 | scf7180000302386:11915-12111 - | -----------------------------------------------------TGGTTTTTGCTGCAAGGCCTTGGTGCACGCAAGGAACAA-GAGAAATACGCAGCAGTAATCGCGAATGCTGATCCCAAATACCTGGATCAAGTGCACGACCTTGTCAATAACCCACCACAAACCAACCCGTACTCAACCCTTAAGCAAGCAATTTTGTCCAAATTTGCCGAATCAGAGATGGTCCGCCTCGACAGACT | |
| droTak1 | scf7180000414611:5988-6184 + | -----------------------------------------------------TGGTTTTTGCTGCAAGGCCTCGGTGCACGCAAAGAGCAA-GAGAAGTACGCAGCAGTAATTGCATACGCTGACCCCAAATACCTCGATCAAGTGCACGATCTTGTCAATAACCCACCGAAAACAAATCCGTACTCCACACTCAAACAAGCAATTTTGTCTAAATTTGCCGAATCAGAGATGGTGCGCCTCGACAGACT | |
| droSim2 | 2l:23018273-23018352 + | A--------------------------------------------------------------------------------------ACGAA-AAAAAGTACAGAGTTGTAATTGACTATAGGAAATTCGAAATCATTGATCAAGGGTTCGATTCTATTAAATAGGA------------------------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
Generated: 05/16/2015 at 08:11 PM