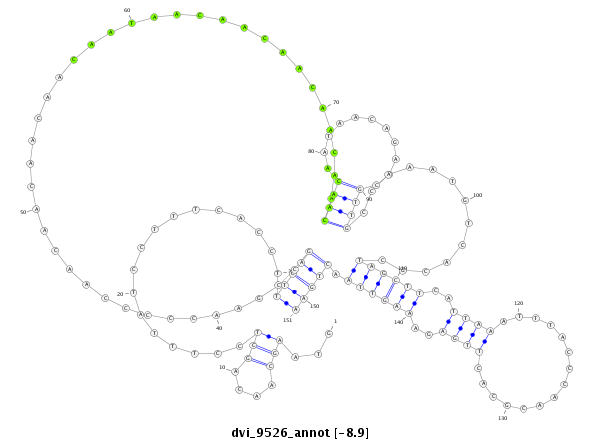
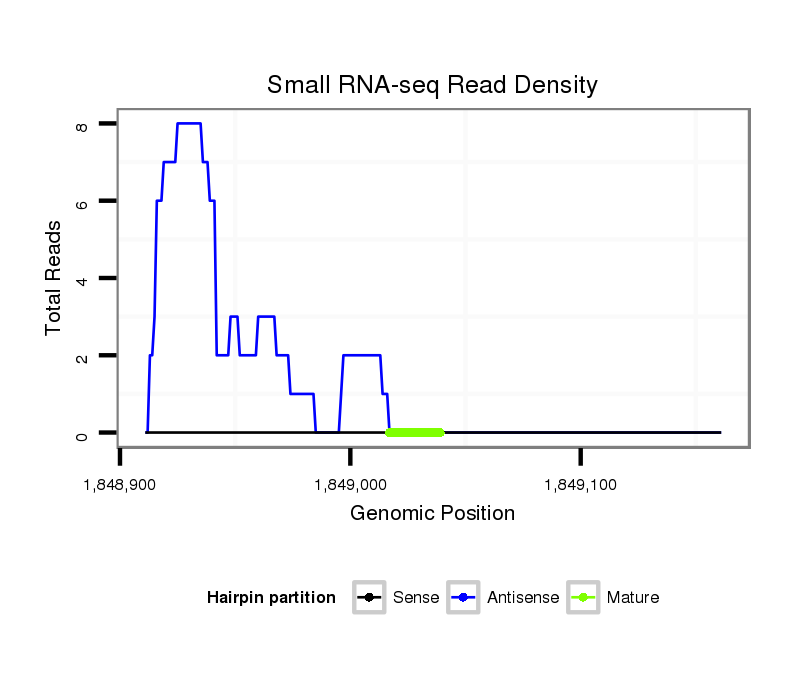
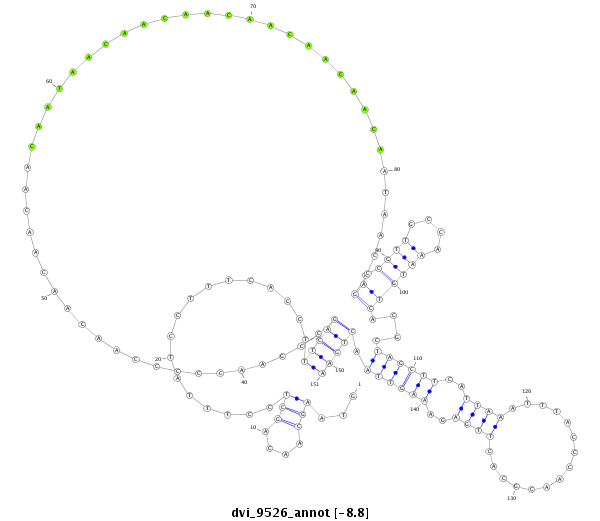
ID:dvi_9526 |
Coordinate:scaffold_12932:1848961-1849111 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.8 | -8.8 | -8.7 |
 |
 |
 |
CDS [Dvir\GJ14875-cds]; exon [dvir_GLEANR_15108:1]; intron [Dvir\GJ14875-in]
| Name | Class | Family | Strand |
| (CAA)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TACCCGTTCTCTATACACGTGGCACTCACTATGAGGTCGGCTTCGATATGGTAAGCAACAGCTCCTTTATCCTTTCACCTTTCAGCCGAACCCCCAACAACAACAACAATAACAACAACAACAACAACAATAACAGACCGTTGCCAAATGTCACGCTAGCTTCATTAAATTTACCCAACGCACTTGAGAAAGTTAACTGAATTTATAAGTTTAGCTTTTCTTTTGATCTAAACTGAAAATTTAACTATTTT **************************************************...(((....))).................(((((.......................................((((...........)))).............((((((..((((...............))))..)))))).)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V047 embryo |
SRR060672 9x160_females_carcasses_total |
SRR060657 140_testes_total |
SRR060674 9x140_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................TCACGCTAGCTTCAAGAAGTT................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................TATGGTAAGCAACGCCTCCG......................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................CTTTCATCAGAACCGCCAAC......................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................CTTTCATCAGAACCGCCAA.......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................CAATAACAACAACAACAACAACA.......................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................AACAACAATAACAACAACA................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
ATGGGCAAGAGATATGTGCACCGTGAGTGATACTCCAGCCGAAGCTATACCATTCGTTGTCGAGGAAATAGGAAAGTGGAAAGTCGGCTTGGGGGTTGTTGTTGTTGTTATTGTTGTTGTTGTTGTTGTTATTGTCTGGCAACGGTTTACAGTGCGATCGAAGTAATTTAAATGGGTTGCGTGAACTCTTTCAATTGACTTAAATATTCAAATCGAAAAGAAAACTAGATTTGACTTTTAAATTGATAAAA
**************************************************...(((....))).................(((((.......................................((((...........)))).............((((((..((((...............))))..)))))).)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060689 160x9_testes_total |
SRR060659 Argentina_testes_total |
V116 male body |
SRR060657 140_testes_total |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
V053 head |
M047 female body |
SRR060670 9_testes_total |
SRR060683 160_testes_total |
GSM1528803 follicle cells |
SRR060687 9_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060666 160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
M027 male body |
SRR060669 160x9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106715 embryo_4-6h |
SRR1106721 embryo_10-12h |
SRR060668 160x9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....CAAGAGATATGTGCACCGTGAGTGAT............................................................................................................................................................................................................................ | 26 | 0 | 1 | 3.00 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CGAGGAAATAGGAAACTGGAAAGTCG..................................................................................................................................................................... | 26 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACCGTGAGTGCTACCCCAGC.................................................................................................................................................................................................................... | 20 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................CCGTGAGTGCTACCCCAGCT................................................................................................................................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AATGTCGGCTTGGGGGTTGTTGTT.................................................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CGGAAATAGGCAAGTGGAAGGT....................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................AACTGGAAAGTCGGCTTGGGGGTTG......................................................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CCAGCCGAAGCTATACCATTCGT.................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GCCGAAGCTATACCATTCGTTGTCGA............................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GAGATATGTTCACCGTGAGTGATACT......................................................................................................................................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GCTTGGGGGTTGTTGTTGTT................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TGTGCACCGTGAGTGATACTCCAGCCG.................................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CCATTCGTTGTCGAGGAAATAGGAA................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CGTTGTCGAGGAAATAGGAAACTGG............................................................................................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GGGCAAGAGATATGTGCACCGTGAGT............................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GAGATATGTGCACCGTGAGTGATACT......................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GCAAGAGATATGTGCACCGTGAGTGAT............................................................................................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................GGCTTGGGGGTTGTTGTT.................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GGGCAAGAGATATGTGCACCGTG.................................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTAGAGGCAATAGCAAAGTG............................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................CCGTGAGTGCTACCCC....................................................................................................................................................................................................................... | 16 | 2 | 16 | 0.31 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...................ACCGTGAGTGCTACCCC....................................................................................................................................................................................................................... | 17 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GGGAATAGTAAAGGGGAAAGT....................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TGGAAAGTGAAAAGTCCGCTTG................................................................................................................................................................ | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GGGAATAGTAAAGAGGAAAGT....................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTCGACGCAATAGCAAAGTG............................................................................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTAGACGAAATAGCAAAGTG............................................................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GTCGGCTTATGGGTTGTAGTT.................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACCGTGAGTGCTACCCCAC..................................................................................................................................................................................................................... | 19 | 3 | 15 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ....................CCGTGAGTGATACCCCCCC.................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GGAAAAAGGCAAGAGGAAAGT....................................................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTCGATGTCTAGGAAATA..................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................AGACGAAGCCATACCATCC.................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACCGTGAGTGCTACTCCCC..................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GGAAATAGTAAAGACGAAAGT....................................................................................................................................................................... | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GTTGTTGTTATTGTTGTTGTTGTTG.............................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACCGTGAGTGCTACCCCT...................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................TAGAGGCAATAGCAAAGTG............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................ACCGGGAGTGATACCCCC...................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12932:1848911-1849161 + | dvi_9526 | TACCCGTT----CTCTATAC----A----CGTGGCACTCACTATGAGGTCGGCTTCGATA--TGGTAAGCAA-------C------------------------------------AGCTCCTTTATCCTTTCACCTTTCAGCCGAACCCCCA------ACAACAACAACAAT------------AACAACA---ACAACAACAACAATAA------------------------------------------C---------------AGAC----------CG--------------T--TGCCAAATGTCACGCTAGCT--------------------------------TCATTAAATTTACCCAACGCACTT---------------------------------------GAGA--------A---------------------------------------------------------------------------AGTTAACTGAATTT----A-----TAAGTTT---------------A--GCTTTTCTTTTGA----------------------------------------------------------------------------------------------TCT-----------------------------------------------------------------------------------------------------------------------------------AAACTGA--------AAATTTAACTATTTT |
| droMoj3 | scaffold_6308:2957640-2957816 - | CTCTCGCC----CATCGTAA----TAAACAACGACG---------------------------------------------------------------------------------------------------------------------------ACAACAACAACAAC------------AACAACA---GCAACA---ACAATAA---------------------------------------TAGC---AA-------------C----------AG--------------C--AGCAAC----AACAACAGCCGTCA---TTCA----------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATAATAAATACATTTGGTATATG------CTCAAATAAATACATCA------AATATTGTGTTGTGATATCCAAATCTAA-------------------------------------------------------------------ATTTA--AATGTT | |
| droGri2 | scaffold_15203:4037552-4037825 + | TACCAGTT----CTTTACAC----C----CGTGGCACTCACTACGAGGTCGGTTTCGATA--TGGTAAGTAT-------TCGTCCTTCGTCCTACGTCCTTC----------------GTCCTTTGTCCTTTGGCCTTTGAA--GTAACTCCA------AAA-CAC-GCTCGTCAAAGTCACGC-CAACCTA---ATTATA------AAAACCAA---------------------------------CCGCTTATT---G------------CGCCAA------TGCCAATAACTGAATTGTACGAAATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGT---------------------------------------------------------------------------------TAAATTTGGGTAAAAACTTTTGAACTTTGTTAAAAGC----------------------A----------ATTGAAAG------------------------------------------------------------GAAAAATGAATTAT-----------------------------------AAGGCTAGCCT | |
| droWil2 | scf2_1100000004590:4427126-4427349 + | ATTTAGTA-----------------------------------------------------------AGCAT-------T-----------------------------------------------------------------------TAACTTTGCATAAAGCAACACC------------ACCAACA---ACAACA------ACAATAACTCAACTCCCACGCTTCTCAG------CAGCAGCAGCAGCTG----------------------------------------------------------------T-----------------------------------------------------------------------------------------------------------------------GCCCTTTACTGCCCCCGTTGGCGTCGCTGCCGCCATCAACGCCACCGCAGCCGCCTCCGCATGACACTTAACGGTATTCATATGGAAACAGAGTTG---CCAATGT---------------------------------------------------------------------------------TGCTTTTG----------------------------------------------------------------------------------------------------------------------------------------------------------------AATTCGA--------GCATTTAAG--CATT | |
| dp5 | XL_group1e:7449421-7449580 - | CAGATGCA----CTTT------------------------------------------------------------------------------------------------------------------------------------------------CAACAACAACAAC------------AACAACA---AAAGGC------AAAGC------------------------------------AAAGAA---------------AAAC----------TA--------------------------------------TTA---G--TG---------------------------------------CACTCGGAGAAAAATGGCATATTAAAAATTAGATTTAAA-AATTAAGAGTGGTGTCAAATTAATAGAATTCATTTTTGGTTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A----AA--------AAACATAATTATTTT | |
| droPer2 | scaffold_2082:636-817 + | CGACTCCG-------------------------------------------------GGA--AAGTAACCAACAGGCAAG-------------------CAAATCCCATATCCCCCAAATTCAAAA------------------CCATCTCCA------TCACCAACAATCAA------------ACCAGCA---ACAGCA------ACAAC------------------------------------ATCAGCGGAC-------------------------AA--------------TTACACAAAATG---------CCCCTGCCCC--TGCCCCTGCTCCAGCCAATGCTCCA----CTAGCTCTACCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC-----------------------------------TCACCTGCAAA | |
| droAna3 | scaffold_13333:878549-878744 - | CTCTACCT----AATTTTTTTCCAT-----------------------------------------------------------------------------------------------------------------------GCAAAACCAAA----ACAAAAA---------------------TAAAAGAAAAAAA------------------------------------------------AACCAA---------------AAAAATATAG------------------------------------------------------------------------------------------------------AAAAAATAAAATAAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAATTTACATTTCGAAGAGC----CTTCTAATTTTTGCCGCACGTTTTCACATTTTTTAATTTTATTTATATACTTTTTTTATTTTTTTAACTT-----------------------------------------------------------------------------------------------------------------------------------ATTCTTT--------ATTTTTAACATTTTT | |
| droBip1 | scf7180000395933:148652-148812 - | CAACAAA-------------------------------------------------------------------------------------------------------------------------------------------AACAC---------CAACAACAACACTA---------GCAACAACA---ACAACAATAACAACAAC----------------------------------------------------------------------------------------------------------AACCATCG---CGTC----------------------------A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTCATAGTCACGTC--------------------------------------AATTTTGTTTGTTGTTGGCCAAAGCTGAGGCTGAGTCTGAGGCCGAGGCTGACTCCGCGTCT---------------------AAAGCTGTGG----CAA--AAGATT | |
| droKik1 | scf7180000302517:776922-777048 + | ACCA------------------------------------------------------------------------------------------------------------------------------------------------------------CGACAACAACAAA------------AACAACCACAACAGCA------ACAAC------------------------------------AGAGAT---------------------GGATGAGAAATT--------------GTAAAAAA-----------------------------------------------------TTT-----A--CACTT---------------------------------------TAGG--------A---------------------------------------------------------------------------AAATCTCTAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTAAGCTTTCGGTGTTTTTAAACCAAAATAA--------A----------AACT | |
| droFic1 | scf7180000454073:2567188-2567379 + | ACCA-----------------------------------------------------------------------------------------------------------------------------------------------------------ACAACAACAACAAG------------AACAACA---ATGGCA------ACAAC------------------------------------AACGGC---AAAGCGGGCTGTAAAA-----------------------------------ATA----TG-----------------C----------------------------------------------CACAAATTGATTGAT-----------TTGCCA----CGGC-------------------------------------------------------------------------------AC------GAAGCTG----AGATCCAAAGA-----C-----------------------------------------------------------------------------GTTTGGCCACGT--TG----------------------------------------CAAACTCGTTTCAGTTTCAATTTCATTTTTAAAGGCA-----TTTTCC------------------------------------------------CCCGCGTTTTTTCC-----------------------------------TCCCCCATCTT | |
| droEle1 | scf7180000491193:278170-278291 + | CTCG-----------------------------------------------------------------------------------------------------------------------------------------------------------GCAGCAGCAACAAA------------AACTACA---ACAACA------ACAAC------------------------------------ATTGCC---TAA-----------------------TT-----------------------------------------------------------------------------------------------------------------------------------------------------GGGCAAGGTCTTTTCTGATCGTAATAAAGTTATTGCCCTCTTCTACTCTCCTTTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T---------------------GGAATTTTAT----CTG--GCTCTT | |
| droRho1 | scf7180000769112:28917-29040 + | CCA------------------------------------------------------------------------------------------------------------------------------------------------------------ACAACAACAGCGA---------------CTTAAGCAACAAC------------------------------------------------AATAGCAAG---G------------CGACAG------TGCC--------------AGCAAAT--------------TG---TGTC----------------------------T--------------------------------------------------------------------------------------------------------------------------------------TGTCATGTAA--AAAAGGTTTTTT-------------------------------------------------------TATAGT-------GCAGAAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGGCTTTTTGTAT-----------------------------------AGATTTTGCTT | |
| droBia1 | scf7180000302422:423794-423921 - | TTTGCCCTTCCGCGTTTTATTAGCT-----------------------------------------------------------------------------------------------------------------------GCAGCTCCA------GCAACAACAATGTGA---------GGCACGCCG---ACAACA---------GC------------------------------AACAACAACACT---------GGTCACAGGC----------GG--------------C--AGCGAC----AGCGCCATCT--------------------------AGCGCTTGA----CTTTGTCCAGCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415868:401543-401726 + | CTCTCAAC----TCTCAAGCGGA----------------------------------------------------------------------------------------------------------------------------AATGGCAGACCAGAGACAACAACAAG------------AATAGCA---ATAATA------ATAACAAG---------------------------------TA---C---------------ATCT----------AG--------------C--AATGGC----AATGGCAGCTCCCG---ACCAGCTTCAGATTCAGTTG---------------------ACACACTG---------------------------------------GGGG--------A---------------------------------------------------------------------------A--AAACTGGGGGC----C-----TAAATTA---GATTGGC---------------------------------------------------------------------------------TTTCATTA----------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATAA--------GACTTTTAT--TACT | |
| droEug1 | scf7180000409533:603406-603550 + | AACCGTA--------------------------------------------------------------------------------------------------------------------------------------------AACCGAAAACCGAAAACAGAAAACCG------------AAAAACA---ATAACA------ATAACAAA---------------------------------CA--GC---A-----AATAACAAA------TAACAAATAAC--------------AGTAAATG---------AACTT-----------------------GACTCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGATTTTCCCCAAATGTAAATT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTATTTTATTCAC-----------------------------------TTACTTGTTTT | |
| dm3 | chr2L:20689431-20689572 - | TAACTTCT----CCACGCAA----ACCGCCTCGACGACCACCACGACGTCGGCATCGGCAGCTGGACACCAC-------C------------------------------------AACACCATAACCACCTCCTCCATCAGCAGCATCAC---------AACCAGCAGCAGCA---------GCAGCAGCA---GCAACAGCAACAACAAC----------------------------------------------------------------------------------------------------------AG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | x:4636279-4636415 + | ACA------------------------------------------------------------------------------------------------------------------------------------------------------------ACAACAATAATGGT------------AACAGAA---GCAACA---------GC------------------------------------AACAACGGCCAA---------------ACATAAACAACAAC-----------------------------AG--------------------------TAA---------------------A--CACTC---------------------------------------GAAG--------A---------------------------------------------------------------------------AAACAACGTAAATA----T-----CAAGTTT---------------TACGCTCTCCCAATTA----------------------------------------------------------------------------------------------CGT-----------------------------------------------------------------------------------------------------------------------------------TACGTTA--------AGATCATATTTTTTT | |
| droSec2 | scaffold_40:58926-59088 + | CAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------AACAATAACAAT------------AACAATA---ACAACAACAATAATAA------------------------------------------C---AA-------------C----------TT--------------------------------------TTT---T--TC---------------------------------------CACGCCCA--------------------------------------------------------------------------CTCTAACGCCCA--CCG--A-------------------------GC------AAAACTG----CCACGTCCATA-----C-----------------------------------------------------------------------------TTTTGAACA-------------------A----------------------GTTTTTAAATTTTTTTTAATTTTA------TTCACCAATAT------------------------------------------------------------------------------------------ACCAGAA--------AAATTATGAAATTTT | |
| droYak3 | X:8074639-8074774 + | TTTGTTCTTGCGTTTTCTTCTGGCT-----------------------------------------------------------------------------------------------------------------------TGACCACCA-----TCAGAGAAATACAAC------------AACAACA---ACAACA------ACAAC------------------------------------AATTGA---------------ACAC----------TG--------------C--GGCGAAATA----TG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAATT----------------ATTATGTTTGCATATCGCCGACT----TG-------------------------------------------------------------------------------------------------------------------------------------------------CTGG-------------------------------------------------------------------GTTT---CATTTT | |
| droEre2 | scaffold_4784:18413974-18414097 - | CAGGAGA-------------------------------------------------------------------------------------------------------------------------------------------GGCAC---------GAGCAGCAGCAGCA---------GCAGCAGCA---GCAACAGCAGGCGCAGC--A-------------------GCAACTGCCGCAGCAACACC----------------------------------------------------------------AA---------------------------------------------------CC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATGCAACTGCAGA-------GCTCAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCAGTTTTTGCAG-----------------------------------TCGGTCTTTTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 05:07 AM