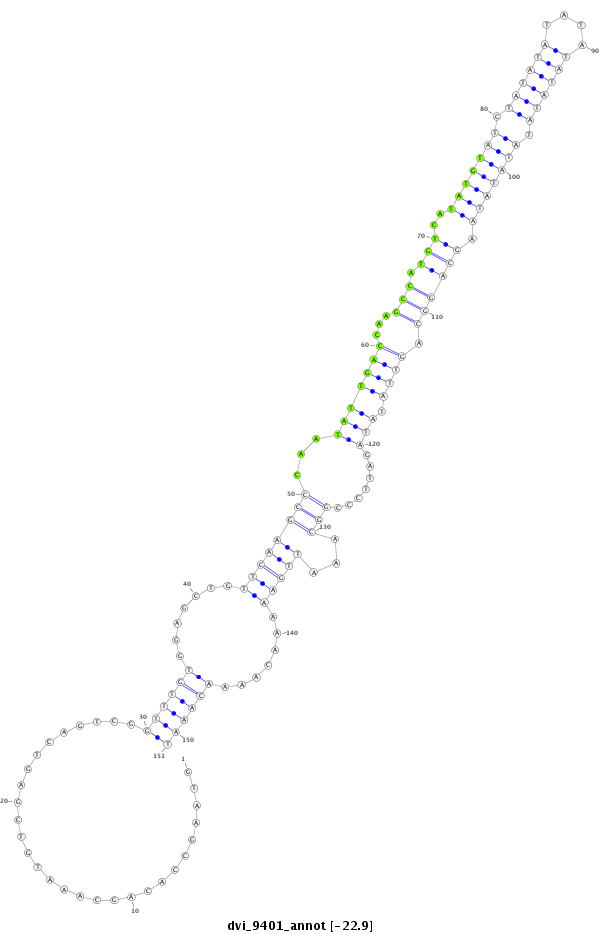
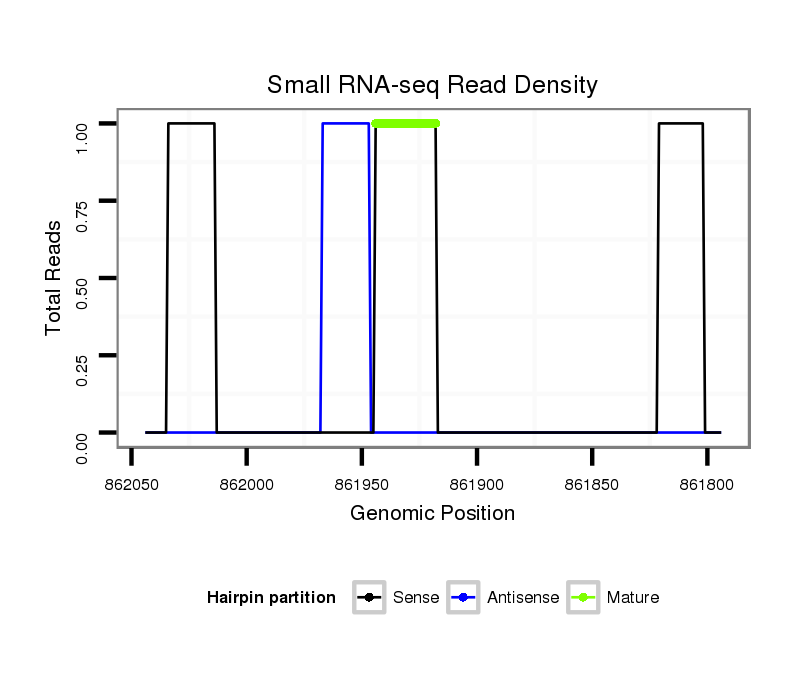
| ATATAG-TACTTATTATTATATACATATAGGTCCATATACATATACATGTATGC-------------------------------------------------------------------------AGA------------------------------------------------------------------AGGCA---------------------------TGCATAT-------------------GTCATGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAGCATTCCCACG-CTCATATATATATACATATATATACATATATATACATATATATACATATATATACATATATATAC-------------------------------------------------------------------------------------------------ATATATATACATATATATACATATATATACAT-ATATA------------------------TACATATATATACATATATATACATATATATACAT--------------------------------------------- | Size | Hit Count | Total Norm | Total | M026
Head | M043
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGC............................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 23.75 | 475 | 0 | 298 | 149 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGC............................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 2.10 | 42 | 0 | 42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................ACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCT.............................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 1.85 | 37 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 36 |
| ............................................................................................................................................................................................................................................................................GACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATG................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.90 | 18 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCAT................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.65 | 13 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| .....................................................................................................................................................................................................................................................................................................................................................................TGCAAAGTGCATGCTCAGCATTCCC.................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.60 | 12 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................TGCAAAGTGCATGCTCAGCATTCCCA................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.40 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| ...............................................................................................................................................................................................................................................................................................................................................................................ATGCTCAGCATTCCCACG-CTC............................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.35 | 7 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................CTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATG................................................................................................................................................................................................................................................................................................................................ | 27 | 20 | 0.30 | 6 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTC............................................................................................................................................................................................................................................................................................................................. | 29 | 20 | 0.30 | 6 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................TGCATGCTCAGCATTCCCACG-CTC............................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGC................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................TAT-------------------GTCATGTATGGTCTGACCACTTG............................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................GCATGCTCAGCATTCCCACG-C............................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGC-------------------------------------------------------------------------AGA------------------------------------------------------------------AGGCA---------------------------TGCATAT-------------------GTCATGTA............................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCA......................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.20 | 4 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCT.............................................................................................................................................................................................................................................................................................................................. | 27 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................TGCAAAGTGCATGCTCAGCATTCC..................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................TATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAG...................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCG.................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................TCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCA.................................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAG...................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................CACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAG........................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................TCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATG................................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................C-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAGCATTCCC.................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................C-------------------------------------------------------------------------AGA------------------------------------------------------------------AGGCA---------------------------TGCATAT-------------------GTCA................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATG................................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGC-------------------------------------------------------------------------AGA------------------------------------------------------------------AGGCA---------------------------TGCATAT-------------------GTCATGT............................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGC.......................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................CTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCA.................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TGCATAT-------------------GTCATGTATGGTCTGACC.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................CCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCA............................................................................................................................................................................................................................................................................................................................ | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................TGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAG...................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................AGTGCATGCTCAGCATTCCCACG-CTC............................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGC................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................ACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCA............................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGC............................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................AT-------------------GTCATGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------............................................................................................................................................................................................................................................................................................................................................. | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................CATGCTCAGCATTCCCACG-CT.............................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................GTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGC.......................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................T-------------------GTCATGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------............................................................................................................................................................................................................................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................CTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGC................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTG.................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................TGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAGCATT....................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAA....................................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................ACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCA............................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................T-------------------GTCATGTATGGTCTGACCACT............................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................ACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGC................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................TGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAA........................................................................................................................................................................................................................................................................................................................................ | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................AAAGTGCATGCTCAGCATTCCCACG-C............................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................AGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAGCATTCCC.................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................TATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAA....................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................CAAAGTGCATGCTCAGCATTCCCACG-................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................GTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCA.................................................................................................................................................................................................................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................TCATGTATGGTCTGACCACTTGAGC-----------------------------------------------------------------------------............................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GC-------------------------------------------------------------------------AGA------------------------------------------------------------------AGGCA---------------------------TGCATAT-------------------GTCATGT............................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGT............................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................TGAGC-----------------------------------------------------------------------------TGCAAAGTGCATGCTCAGCA......................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................TGCAAAGTGCATGCTCAGCATTCCCACG-................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................TGGTCTGACCACTTGAGC-----------------------------------------------------------------------------TGCAAAGTG.................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................TGCAAAGTGCATGCTCAGCATTCCCACA-................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |