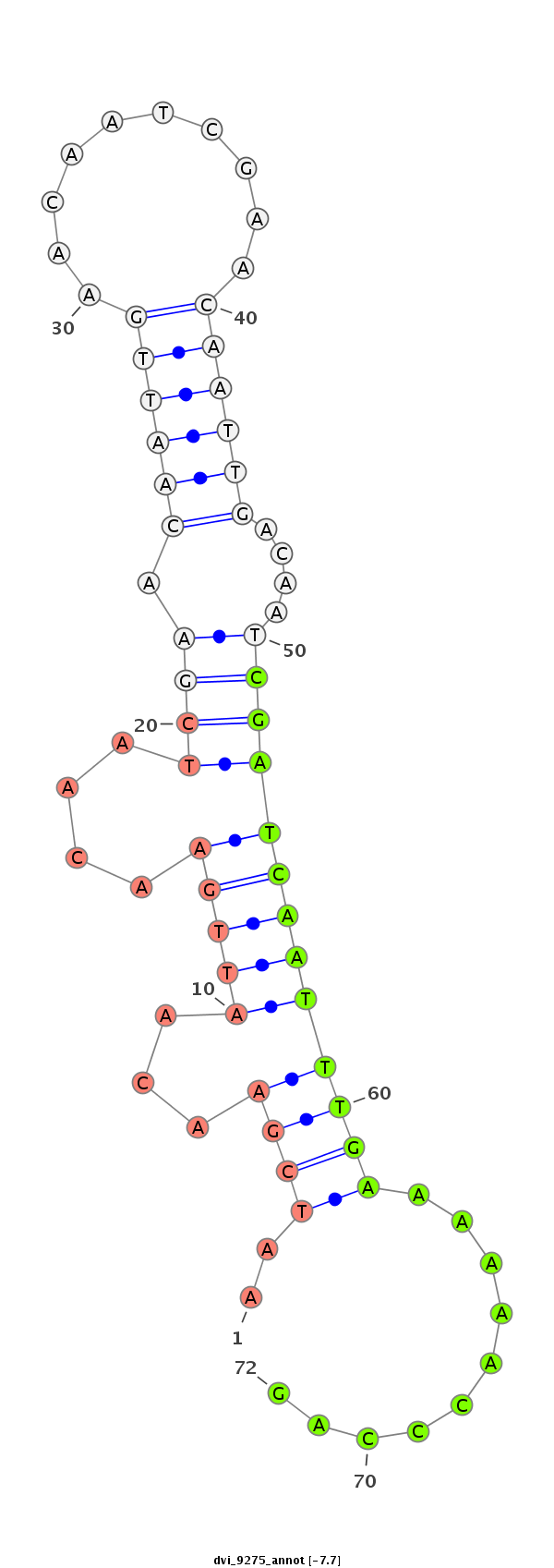

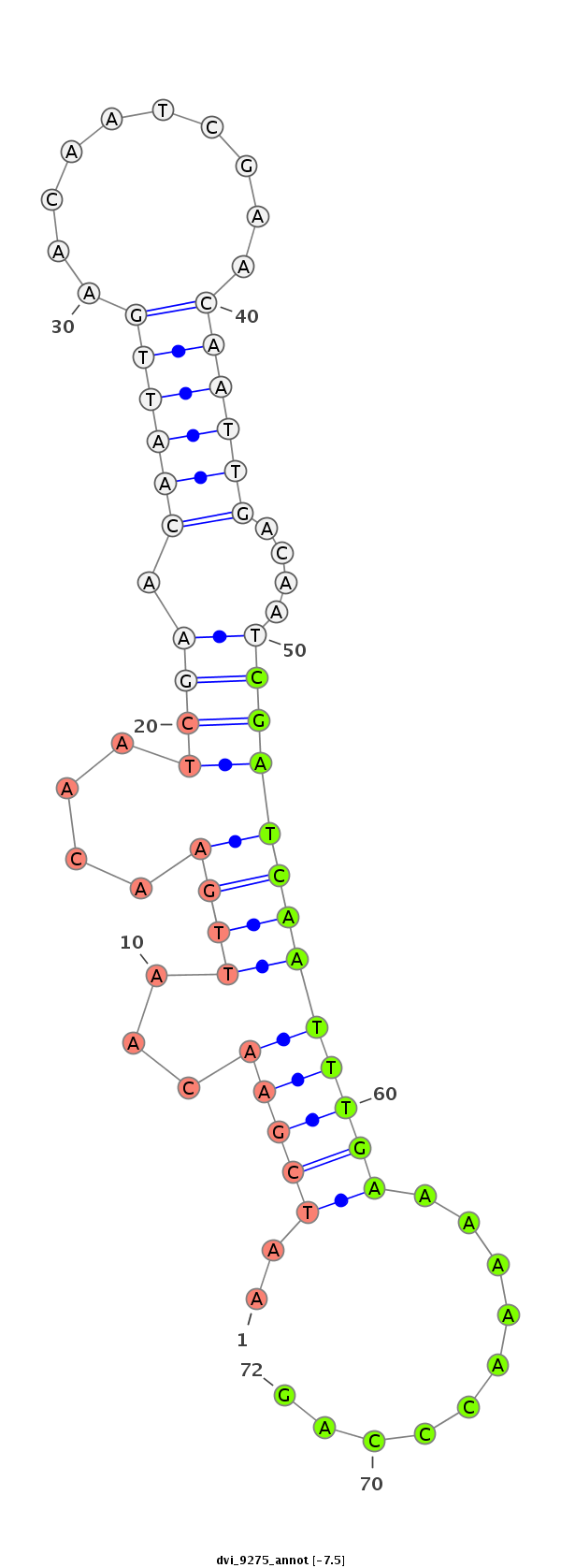
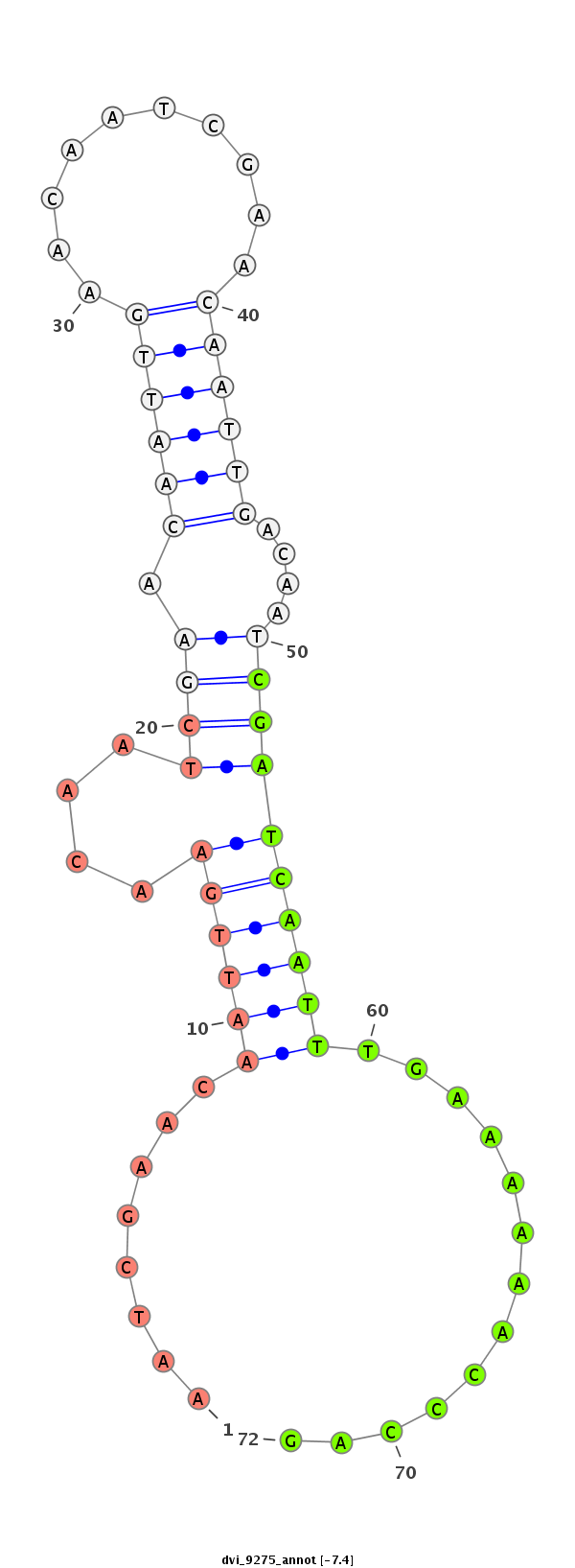
ID:dvi_9275 |
Coordinate:scaffold_12928:7458481-7458631 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
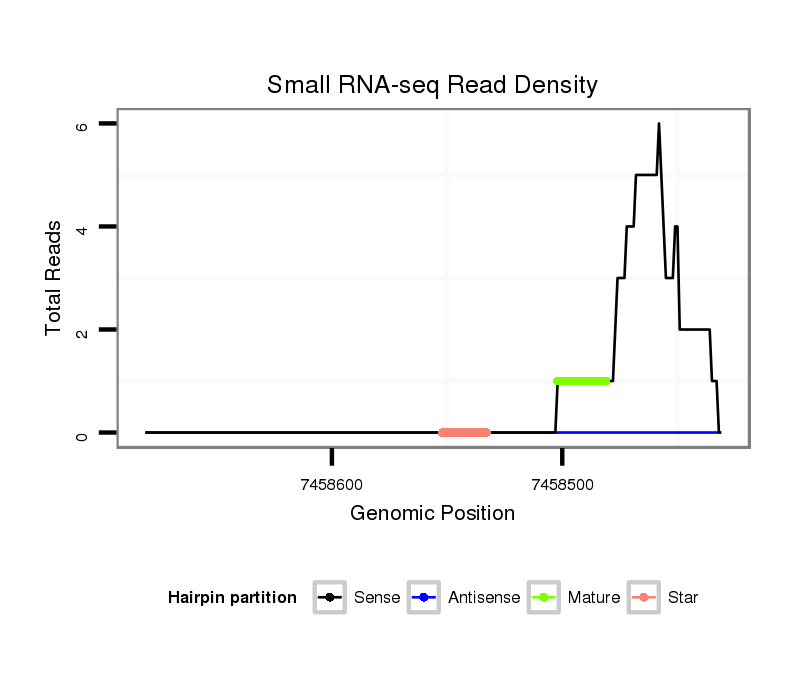
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -7.5 | -7.4 | -7.3 |
 |
 |
 |
exon [dvir_GLEANR_16871:2]; CDS [Dvir\GJ16365-cds]; intron [Dvir\GJ16365-in]
| Name | Class | Family | Strand |
| (ATTG)n | Simple_repeat | Simple_repeat | + |
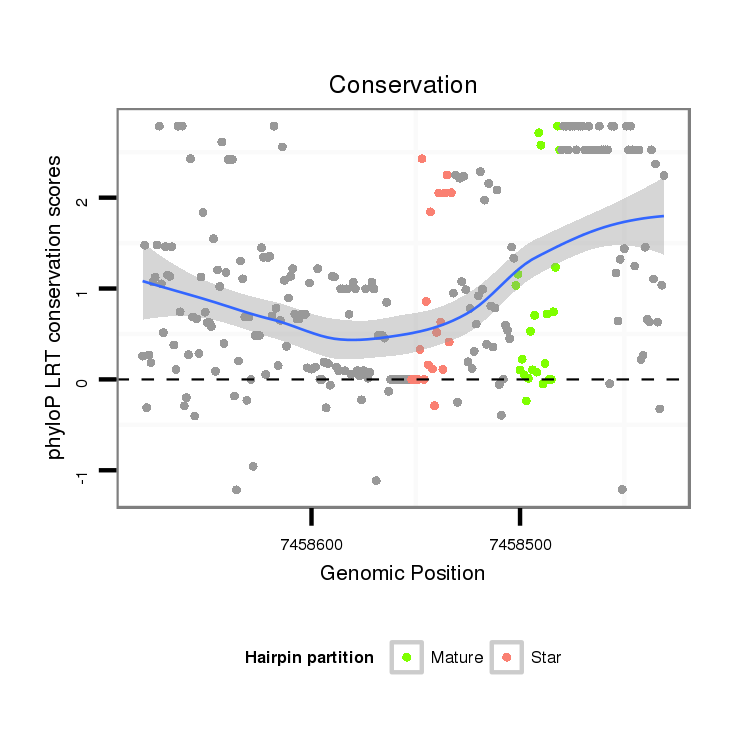
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTGCCGCTTTGTTTTCTTCTTCTTTTCTCTCTTATTTTTCTTTTTGATTTTTCGAAATTCGTTTTGTTCATCATCGTTATCGTTATATTGTCATTGCACACACATCACGCACCAATCGAACAATCGAACAATCGAACAATTGAACAATCGAACAATTGAACAATCGAACAATTGACAATCGATCAATTTGAAAAAACCCAGGATCTTACATTCCAGGGCTCGGGCTATCCAATGTCACCATTGGGTGTCGC *********************************************************************************************************************************..((((...(((((....((((.((((((..........))))))....)))))))))))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M047 female body |
SRR060672 9x160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
M061 embryo |
SRR060656 9x160_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060671 9x160_males_carcasses_total |
V116 male body |
SRR060659 Argentina_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
M028 head |
SRR060665 9_females_carcasses_total |
SRR060675 140x9_ovaries_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TTACATTCCAGGGCTCGGGCT......................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CGATCAATTTGAAAAAACCCAG.................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TGCTTTTTGATTCTTCGA.................................................................................................................................................................................................... | 18 | 2 | 20 | 1.00 | 20 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 4 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| .....................................................................................................................................................................................................................CAGGGCTCGGGCTATCCAA................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ATTCCAGGGCTCGGGCTATCCAA................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CTTACATTCCAGGGCTCGGGC.......................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AATGTCACCATTGGGTGTC.. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GCTATCCAATGTCACCATTGGGT..... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GATCTTACATTCCAGGGCTCGGG........................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTGCTTTTTGATTCTTCGA.................................................................................................................................................................................................... | 19 | 2 | 20 | 0.70 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................AATCGAACAATTGAACAATC...................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AATCGAACAATTGAACAATC...................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTTCTTTTTGATTCTTCG..................................................................................................................................................................................................... | 18 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTGCTTTTTGATTTTTCGAT................................................................................................................................................................................................... | 20 | 2 | 17 | 0.24 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTTCTTTTTGATTCTTCGAT................................................................................................................................................................................................... | 20 | 2 | 12 | 0.17 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................CTTTTTGATTCTTCGA.................................................................................................................................................................................................... | 16 | 1 | 19 | 0.16 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................ATCAAACAATCGAGCAATCGT.................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TCCTTTTTGATTCTTCGA.................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................ATTTCTTTTTGATTCTTCG..................................................................................................................................................................................................... | 19 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TCTCGGACCAATCGAACCA................................................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CATCTAGGACCAATCGAAC.................................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................CTTTTTGATTCTTCGATAT................................................................................................................................................................................................. | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................ACTGATCGGGCTATCCAAT.................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................TTGCTTTTTGATTTTTCGATGT................................................................................................................................................................................................. | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................ATTGCTTTTTGATTCTTCGAA................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTGCTTTTTGATTCTTCG..................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................GATGAATTTGAAAAAGCGCA................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................GCTTTTTGATTCTTCGAA................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AACGGCGAAACAAAAGAAGAAGAAAAGAGAGAATAAAAAGAAAAACTAAAAAGCTTTAAGCAAAACAAGTAGTAGCAATAGCAATATAACAGTAACGTGTGTGTAGTGCGTGGTTAGCTTGTTAGCTTGTTAGCTTGTTAACTTGTTAGCTTGTTAACTTGTTAGCTTGTTAACTGTTAGCTAGTTAAACTTTTTTGGGTCCTAGAATGTAAGGTCCCGAGCCCGATAGGTTACAGTGGTAACCCACAGCG
**************************************************..((((...(((((....((((.((((((..........))))))....)))))))))))))..........********************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060666 160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060665 9_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060668 160x9_males_carcasses_total |
SRR060678 9x140_testes_total |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................AGAGATTAAAAAGAAAAAC............................................................................................................................................................................................................. | 19 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................TCAGCAAAGCAAGTAGTAG................................................................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TTAAGCAAGACATGTAGTAGG............................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GCGTGGTTAGCTGGTGAG.............................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............AAGAAGGAGAAAGGAGAGAA.......................................................................................................................................................................................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............AAAGCAGAAGAAAGGAGAG............................................................................................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................AACAAGCAGTGGCAATAG.......................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GCGTTGTTAGCTTGTCAG.............................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GCAAAGGAAGTAGTAGCTATA........................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................AAGGGCCCGAGGTCGATAG...................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................GTTAACTGTTAGCCCGTTA................................................................ | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGCTTGTTAACTCGATAGC..................................................................................... | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TAGTTTGTTAGCTCGTTAGCA.................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AGCTTGTTAACTCGATAGC..................................................................................................... | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGACCAAGTAGTAGCATTA........................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:7458431-7458681 - | dvi_9275 | TTGCCGCTTTGTTT--TCTTCTTCTT--TTCTCTCTTA----TTTT------------TCT-------T-TTT------GATTTTTCGAAA---------TTCGTTTTGT-TCA-----------------------------------------------T---------CATCGTTA------TCGTTATATTGTCAT--------------TGCA-CACACATCACG---CACCAATCGAACAATCGAACAATCGAA-----CAATTG-AACAA---------TCGAACAATTGAAC------------------------AATCGAAC-----------------------------------AATTG-------------------------------------------------------------------------------AC--A-ATCGA------T--CA-------ATTTGAAAAAACCCAGGATCTTACATTCCAGGGCTCGGG---CTATCCAATGTCACCATTGG---GTGTCGC |
| droMoj3 | scaffold_6473:2582521-2582749 + | TTATCGCTTTTTTT--C-TTTTCTC--------TCCATTCTATTT------------TTCT-------T-TTTTTTTTC--------CAAATTTTTAT-TTTGAATTCGT-T----------------ACA-----------------------------AT---------CATCGTTATA----TCGTTA--TTGTCAT--------------TGCA-CATACACCACG---CACGAATCGA----------------------------------------------------------------------------------------------------------------------------------ACAATCGACCAACAATCG--------------AACCAT------------------------------------CGA-----A-ATCGT------T--CAATTTGAAA-ACCAACC---AAAGGATCTTACATTCCAGGGCTCGGG---CTATCCAATGTCACCGTTGG---GTGTGGC | |
| droGri2 | scaffold_15081:3239528-3239799 + | TTGTCGCTTTGTTT--TCTTCTCCT---------------------CCTCTTAATTTTTTT-------T-TTATTTTCT--------------------TTTTATTTTCT-TTCTAATC-----G------------------------TTTTCATCATTAT---CATCATCATCGTTA------TCGTTA--TTGCCATCATCTAATAATCAATGCACCACACACTACG---CACT-------------------------------------------------------------GC---------------------CAACAACAAAAAA------------ACCGAATCGTTATCGATAACCGATAT-CCGATA---------------------A-------------------------------TTCGATGAATT------GAAT--G-A---A------A-------------ACGA-AC---ACAGGATCTTACATTCCAGGGCTCGGGAGGGTATCCGATGTCACCATTGA---GTGGCGC | |
| droWil2 | scf2_1100000004963:2952887-2953086 - | TTTTTTTTTGTTTT--TCTTTTTGGT------T-T----------------------TTAT-------T-TTGTTTTTC--------------------TTTTATGTTT------AATT-----T----------------TT----------CAT--------------------------------------------------------AACGCGCC-CACGCTGCTGATTACT----GC--------------AAA-----C-----CACAAA---------TCAATCCAC---------------------------------------------------------------------------------------AACAAATCG--------------AA-------------------------ATCG---------ATCGAAAT--G-GTGAA------A-----------ATCTCAA-T---GCAGGATCTTACATTCCAGGGCTCGGGTGGTTATCCCATGTCACCACTGG---GTGTGGC | |
| dp5 | XL_group1e:6583236-6583475 - | dps_2720 | ATGC---TTTTTGC--TTGTTTCTTTTCTGCTTTCTTT----TCTTCCTTTTAC-------------------T-----GATTTCT---------TTT-GTTTATGTTGTTTTGCACTCTACTCGAACAACCAA---------------------------------------------------------------------------------------------------------------------------CAA-----CAAC----------------------CAACAACA------------------ACAACAACAACAACAAA---CAACAACTAAATGCATC----------------------------------------------------ATCATCAACATCCTACAACAT------------------------CC--T-ACGGGCATCCGAACCG-------CACTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---TTATGCGATGTCACCATCGATGGG------ |
| droPer2 | scaffold_21:1595482-1595709 - | ATGC---TTTTTGC--TTGTTTCTTTTATGCTTTCTTT----TCTTCCTTTTAC-------------------T-----GATTTCT---------TTC-GTTTATGTTGTTTTGCACTCTACTCGAACAAC------------------------------------------------------------------------------------------------------------------------------CAA-----CAAC----------------------CAACAACA--------------A---ACAACAACAACAACAAA-------------ATGCATC----------------------------------------------------ATCATCAACATCCTACAACAT------------------------CC--T-ACGGGCATCCGAACCG-------CACTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---TTATGCGATGTCACCATCGATGGG------ | |
| droAna3 | scaffold_13117:2160382-2160639 - | TTT-CGCTT--TGCTTT-CTTTATTT-----TTTCTTT----TCTTTTTT--------TCA------------TTAT--CCTT------------TTGGCCCGATTTTGT-TTGCACTC-----CACCACC-----------------------------AC---CACCACCA------CGTCCAACAACA--CCACCAC---------------------------------------------------------CA-----------------------------------C------------------------AACAACAACAACA-----------------------------------AACCAACCAACCAACCAACAACTACAAC----AAA-CAA--AATTCA-------------CATCATCCAACGGATA------TAAC--G-ATGGCCATC--A--TC-------CACTGAA-A---ACAGGATCTTACATTCCAGGGCTCGGG---CTATACCATGTCGCCGCTGG---GCGTCGC | |
| droBip1 | scf7180000395304:31742-32002 + | TTT-CGCTT--TGCTTT-CTTTATG--------TCTTT--------------------TCTTTTTTGAT-TTC------AATT------------CTGCCCCGATTTTGT-TTGCACC----TCCACCAAC-----------------------------ACCACCACCACCA------CGTCC------A--CGACC------------------------------------------------------------------------------ATCATGTCCACCAACCA-------------CCACAACAACAACAACAACAACAACA-----------------------------------AACAA-CAAACA---------------------AA-CAAACAATCCA-------------CATCATTCGATGGATA------AAAC--G-ATGGGAATC--A--TC-------AACTGAA-A---ACAGGATCTTACATTCCAGGGCTCGGG---CTATACCATGTCGCCGCTGG---GCGTTGC | |
| droKik1 | scf7180000302398:486762-486956 - | ATGCCGCTT-TTGC--TCCTTTATTTC----GTTCTTT--------------------TCT-------TTTTCTT---CCAT-------------TAC-GTTGATTTCGT-TTGCACTC-----GATCAATC----AATCAATCAATCGAACA--------C-----------------------------------------------------------------------------------------------CAA-----T-----CACCAA---------TCAATCAACCAAAC------------------------CAACC--------------------------------------------------------------------------------------------------------------------------AC--GAATGGG------A--TC-------TTCTGAACCA--ACAGGATCTTACATTCCAGGGCTCGGG---CTACACACTGTCGCCATTGG---GCGC--- | |
| droFic1 | scf7180000454077:2060054-2060324 + | ATGCCGCTTTTTGC--TCGTTTATTT-----TT-------------------------TCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACT--ACTCAACCAACCAACCAATCAATCAATCAATCAAACGAAC--CAC--------------------------------------------------------------------------------------------CAA-----CCACCACACGAA---------TCAATCAACAACAC---------------------CACCAA---CCAACCA-----A----------CT---ACAACAACAAATCA-CCAATCA------------ACCAA--CAACCAACTCATCAT-------------------T---------AAACCGAAACGTG-ATGGG------A--TC-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTGG---GCGTTTC | |
| droEle1 | scf7180000491006:1443701-1443968 - | ATGCTGCTTTTTGC--TTGTTTATTT------TTCTTT----T--------------TTCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAATC----AATCGTTCGTTCGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAC-----CAACCACACCAATCAAATCAATCAATCAACAACC--------------A---ACACCACCAACAACAAA-------------CC------------------AACTA-CCAATCA------------ACCAC--CAACCAACTCATCA------------------------------AAACCGAAAC--G-ATGGG------A--TC-------TTCTCAATC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTGG---GCGTCTC | |
| droRho1 | scf7180000763611:274366-274604 + | AAGCCGCTTTTTGC--TTGTTTTATT------TTCTTT--------------------TCT-------T-TTCTTTTGC--------------------ATTGATTTTGC-TTGCACTC-----AACCA--------ATCAATCGATTGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAC-----CAACCACACCAA---------TCAACCAACAACAC---------------------CACCAACAACA-----------------------------------AACCA-CCAATA---------------------AATCAACTCATCAA-----------------------------AAACCAAAAC--G-ATGGG------T--TC-------TTCTGAATC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAACATGTCGCCGCTGG---GCGTCTC | |
| droBia1 | scf7180000301623:228744-229024 + | ATTCCGCCTTTGGC--TCGTTTATTT------T-------------------------TCT-------T-TTATTTTGC--------------------GTTGATTTTGC-TTGCACTC-----AATC--------AATCAATCGATCGATCAACTCAACACCACCAC-----------------------------------------------------------------------------------------CAC-----CAACCACACCAA---------TCAATCAACAACACCAACTACAACAACAACAACAACAACAACAACA----------ACTA-------CTACAACAACAACAAACCA-CCAATC---------------------AACCAACTCATCAA-------------------CC--------AAACCGAATC--G-ATGGG------A--CG-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTGG---GCGTCTC | |
| droTak1 | scf7180000415141:16362-16617 - | TTGCCGCTTTTTGC--TCGTTTATTT------TTCTTT----T--------------CTCT-------T-TTATTTTGC--------------------ATTGATTTTGT-TTGCACTC-----AACCAATC----AATCAATCGATCGATCAAATCAACACCACCAA-----------------------------------C-----------------------------------------------------CAACCAACCAACCACACCAA---------TCAATCAACAACAA---------------CACCAACAACAACAACA-----------------------------------AAACG-AAAACC---------------------AACCAACTCATCAA-----------------------------AAACCGAATC--G-ATGGG------A--TC-------TT----------ACAGGATCTTACATTCCAGGGCTCGGG---ATACAACATGTCGCCGCTGG---GCGTCTC | |
| droEug1 | scf7180000409064:24839-25053 - | ATTCCGCTTTTTGC--TCGTTTATTT------T-------------------------T-------------CTTTTGC--------------------GT----TTTTT-TTGCATTC-----AACCA------------ATCGATCGATTAAATCAAAAC-----------------------------------------------------------------------------------------------CAC-----CAATTACACCAA---------CCAACCAACAACAC------------------------C---AACA-----------------------------------AACCA-CCAATC---------------------AACAAACTCATCTA-----------------------------AAATCGAAAC--G-ATGGG------A--TC-------TTTTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTGG---GCGTCTC | |
| dm3 | chrX:7947278-7947520 - | ATGCTGCTTTTTGC--TCGTTTTATT------T-------------------------TCT-------TTTTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAACCAACCAATCAATCGATCGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAA-----CAACCACACCAA---------TCAATCAACAACAC---------------------CAACAACAACA-----------------------------------AACCA-CAAATC---------------------AACCAAATCATCAA-----------------------------AAACCGAAAC--G-ATGGG------T--CA-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTAG---GCGTCTC | |
| droSim2 | x:7519836-7520070 - | ATGCTGCTTTTTGC--TCGTTTTATT------T-------------------------TCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAACC----AATCAATCGATCGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAA-----CAACCACACCAA---------TCAATCAACAACAC------------------------CAACAACA-----------------------------------AACCA-CAAATC---------------------AACCAAATCATCAA-----------------------------AAACCGAAAC--G-ATGGG------T--CA-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTAG---GCGTCTC | |
| droSec2 | scaffold_44:31602-31835 - | ATGCTGCTT-TTGC--TCGTTTTATT------T-------------------------TCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAACC----AATCAATCGACCGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAA-----CAACCACACCAA---------TCAATCAACAACAC------------------------CAACAACA-----------------------------------AACCA-CAAATC---------------------AACCAAATCATCAA-----------------------------AAACCGAAAC--G-ATGGG------T--CA-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTAG---GCGTCTC | |
| droYak3 | X:13364810-13365059 + | ATGCTGCTTTTTGC--TCGTTTATTT------T-------------------------TCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAACC----AATCAATCGATCGATCAAATCAACACCAACAA-----------------------------------------------------------------------------------------CAA-----CCACAACACCAA---------TCAATCAACAACAC------------------------CAACAACA-----------------------------------AACCA-CAAATCA------------ACCAAATCAACCAAATCATCAA-----------------------------AAACCGAAAC--G-ATGGG------A--CA-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTTG---GCGTCTC | |
| droEre2 | scaffold_4690:16856168-16856406 - | ATGCTGCTTTTTGC--TCGTTTATTT------T-------------------------TCT-------T-TTCTTTTGC--------------------GTTGATTTTGT-TTGCACTC-----AACCAACCAATAAATCAATCGATCGATCAAATCAACAC-----------------------------------------------------------------------------------------------CAA-----CAACCACACCAA---------TCAATCAACAACAC------------------------CAACAACA-----------------------------------AACCA-CAAATC---------------------AACCAAATCATCAA-----------------------------AAACCGAAAC--G-ATGGG------A--CA-------TTCTGAACC---ACAGGATCTTACATTCCAGGGCTCGGG---ATACAATATGTCGCCGCTAG---GCGTCTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 09:56 PM