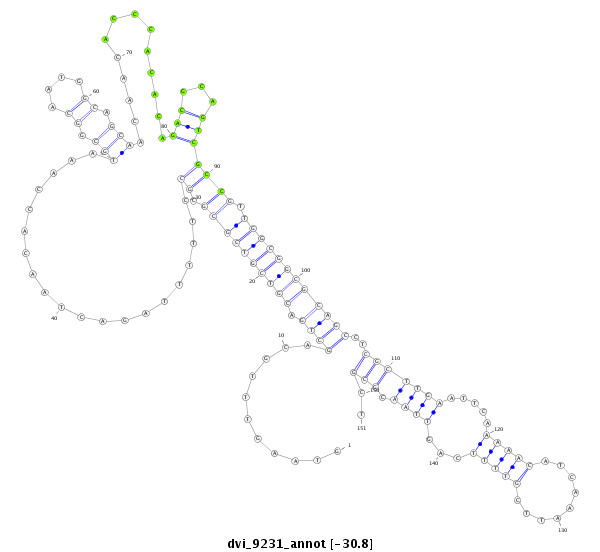
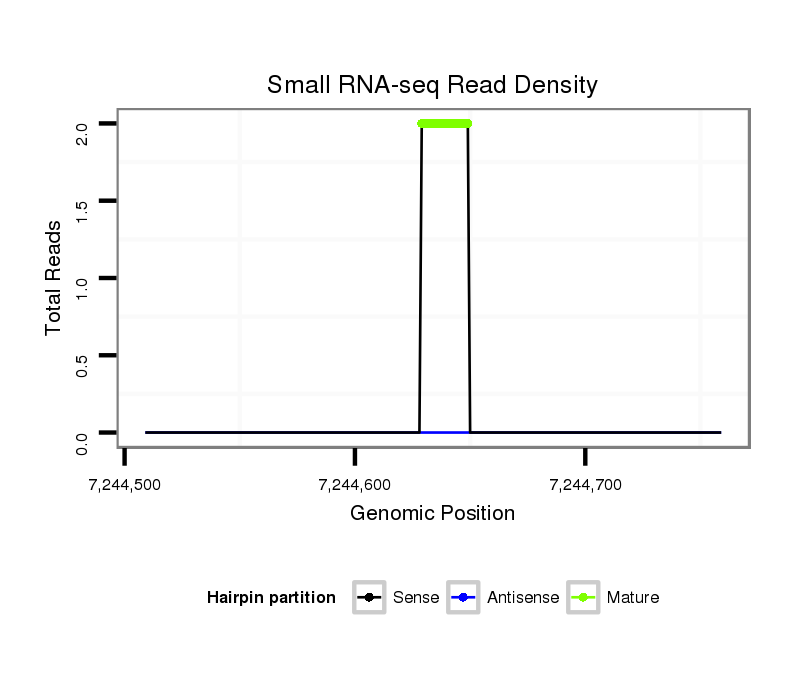
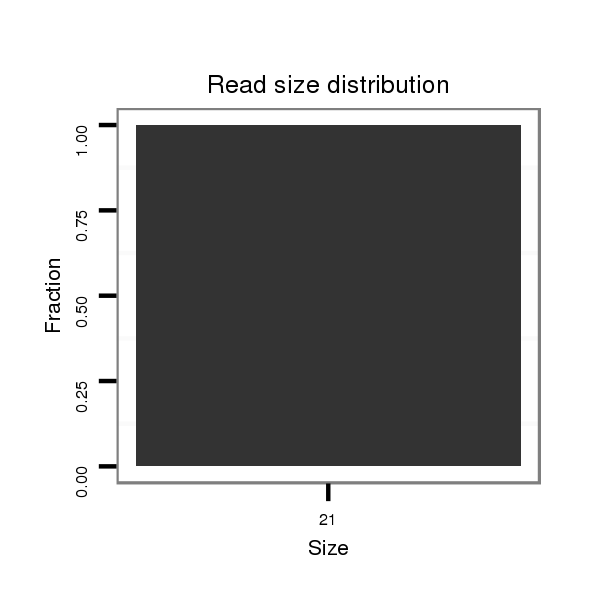
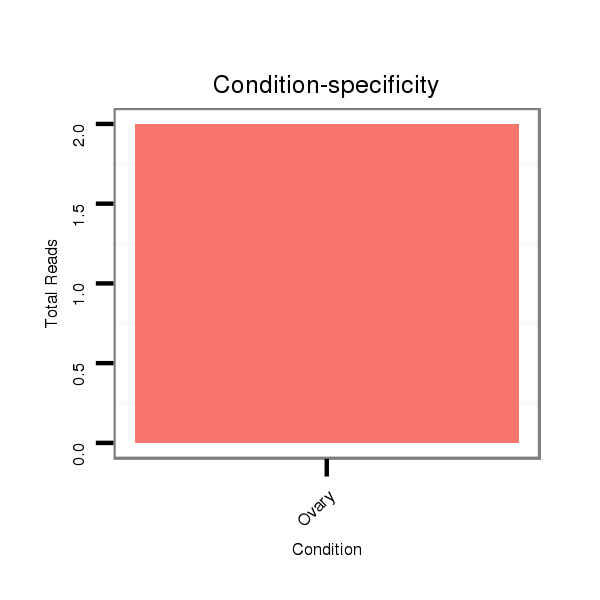
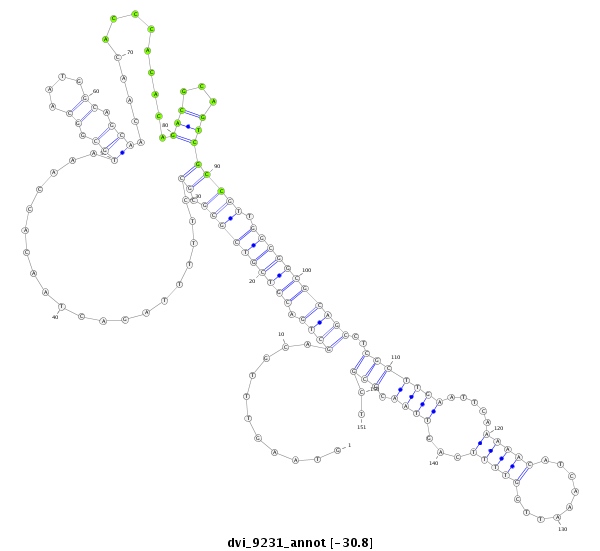
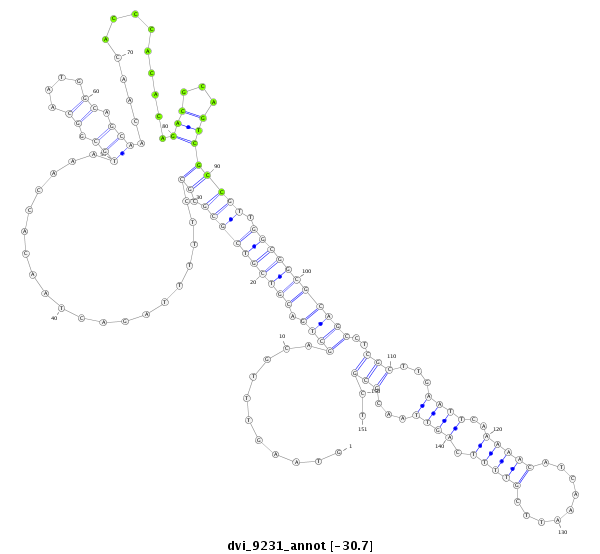
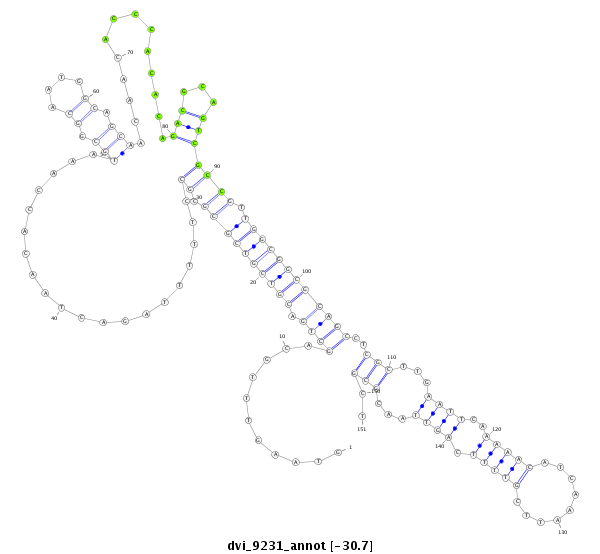
| CAGCAGCAACAACTGCAACAGCA-ACAACAG------CAA---CTG-------------------------------------------------------------------CAG----CAGCAACTCGAG-----------------------------------------------------------------------CAGCAGC--------------------------------------------------------------------------TAGAGCAACA---GGAGCAGCAACAACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCAGCAGCCGATGGTGCAG------------------------------------------C----------------------------------------------------------------------------------------------------------------------------------- | Size | Hit Count | Total Norm | Total | M044
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| ......................................AA---CTG-------------------------------------------------------------------CAG----CAGCAACTCGAG-----------------------------------------------------------------------CA............................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................................G----CAGCAACTCGAG-----------------------------------------------------------------------CAGCAGC--------------------------------------------------------------------------TAGAGCA...................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................GCAGC--------------------------------------------------------------------------TAGAGCAACA---GGAG............................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................ACAG------CAA---CTG-------------------------------------------------------------------CAG----CAGCAACTCGAG-----------------------------------------------------------------------C............................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................G------CAA---CTG-------------------------------------------------------------------CAG----CAGCAACTCG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................ACAACAG------CAA---CTG-------------------------------------------------------------------CAG----CAGC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................CGAG-----------------------------------------------------------------------CAGCAGC--------------------------------------------------------------------------TAGAGCAACA---GG.............................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................GCAACA---GGAGCAGCAACAACAGCAAC------------................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................G-----------------------------------------------------------------------CAGCAGC--------------------------------------------------------------------------TAGAGCAACA---GGA............................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......AACAACTGCAACAGCA-ACAACAG------C................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................G------CAA---CTG-------------------------------------------------------------------CAG----CAGCAACTCGAG-----------------------------------------------------------------------.............................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GCAGC--------------------------------------------------------------------------TAGAGCAACA---GGAGC........................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........ACTGCAACAGCA-ACAACAG------C................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCAGCA........................................................................................................................................................................................... | 25 | 20 | 0.20 | 4 | 0 | 1 | 0 | 0 | 0 | 3 |
| ....AGCAACAACTGCAACAGCA-................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................AAC------------AG-----CA------------------------------------------------GCAGCAGCAGCAGC......................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................ACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCAGC............................................................................................................................................................................................ | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................CAAC------------AG-----CA------------------------------------------------GCAGCAGCAGCA........................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................AGCAACAACAGCAAC------------AG-----CA------------------------------------------------GC..................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCA.............................................................................................................................................................................................. | 22 | 20 | 0.10 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................CAAC------------AG-----CA------------------------------------------------GCAGCAGCAGCAGC......................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................CAACAACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGC............................................................................................................................................................................................... | 25 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................AACAACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGC............................................................................................................................................................................................... | 24 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................AGCAACAACAGCAAC------------AG-----CA------------------------------------------------GCAGC.................................................................................................................................................................................................. | 24 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................................................................................................GCAGCAACAACAGCAAC------------AG-----CA------------------------------------------------GCA.................................................................................................................................................................................................... | 24 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................CAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCA.............................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................................................................................................................AGCAGCAACAACAGCAAC------------AG-----C........................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................ACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGC............................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GCAGCAACAACAGCAAC------------AG-----C........................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AACAGCAAC------------AG-----CA------------------------------------------------GCAGCAGCAGC............................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................AGCAGCAACAACAGCAAC------------AG-----CA------------------------------------------------....................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................ACAACAGCAAC------------AG-----CA------------------------------------------------GCAGC.................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................AACAACAGCAAC------------AG-----CA------------------------------------------------GC..................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |