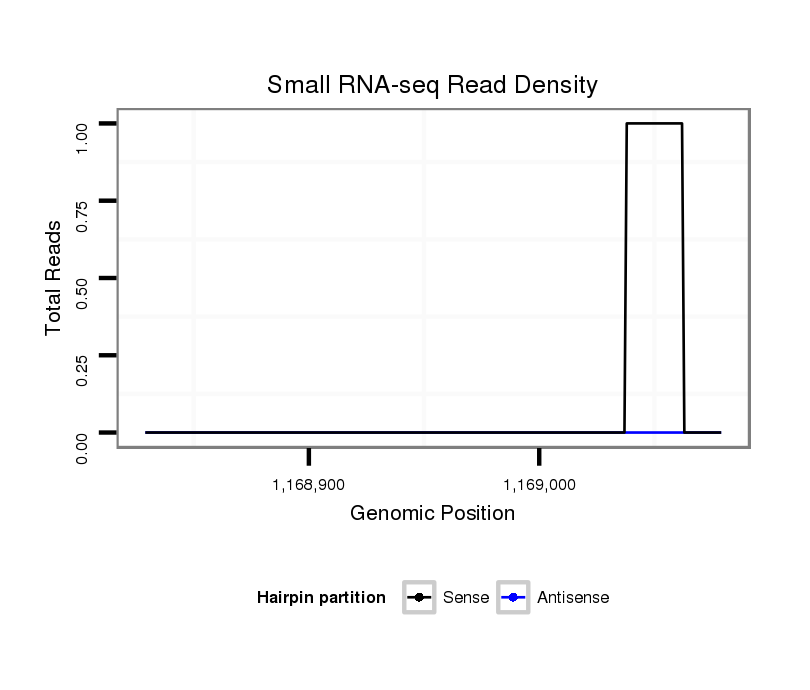
| CAGCAGC--A--ACAACAGCAGCAGCAGCAACAACAACAGCAAC---------AACAGCAGCAGCAAC------A-------AC----AACAACAG---CA-----GCA---------G------CAGCAGCAACAACAA------C-----------AACAGA-------------------------------------------------------------------------------------------------------------------------ATCCTCCGCCTCGCAGATCCCCAAATGATTTCATCTTTGGC---------CG-----------------------------------------------------------------------------------------------------------------------------------------------------CTAC-----------------ATCGGGGAG-----------------------------------------------GGCAGCTTCAGCAT---------------------------------------------------------------------------------------------------------------------------------------------------------G | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ......................................................................................................................................CAACAA------C-----------AACAGA-------------------------------------------------------------------------------------------------------------------------ATCCTCCGCCTCGCA............................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TCCTCCGCCTCGCAGATCCC......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................CCGCCTCGCAGATCCCCAAA..................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................A-------------------------------------------------------------------------------------------------------------------------ATCCTCCGCCTCGCAGATCCC......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................CTTTGGC---------CG-----------------------------------------------------------------------------------------------------------------------------------------------------CTAC-----------------ATCGGG.......................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GA-------------------------------------------------------------------------------------------------------------------------ATCCTCCGCCTCGCAGATCCCC........................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TTCATCTTTGGC---------CG-----------------------------------------------------------------------------------------------------------------------------------------------------CTAC-----------------ATCGGG.......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................GGC---------CG-----------------------------------------------------------------------------------------------------------------------------------------------------CTAC-----------------ATCGGGGAA-----------------------------------------------........................................................................................................................................................................ | 18 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....AGC--A--ACAACAGCAGCAGCAGC.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................ACAGCAGCAGCAAC------A-------AC----AACAACAG---C....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GCAGCAGCAAC------A-------AC----AACAAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................AAC------A-------AC----AACAACAG---CA-----GCA---------G------CAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...........................GCAACAACAACAGCAAC---------AACAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CAACAACAGCAAC---------AACAGCAGCAGC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CAACAACAGCAAC---------AACAGCAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AACAACAACAGCAAC---------AACAGCAGCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CAAC------A-------AC----AACAACAG---CA-----GCA---------G------CAGCA......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CAGCAGCAAC------A-------AC----AACAACAG---........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................AGCAAC------A-------AC----AACAACAG---CA-----GCA---------G------C............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CAGCAGCAAC------A-------AC----AACAACAG---C....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................GCAAC------A-------AC----AACAACAG---CA-----GCA---------G------.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...CAGC--A--ACAACAGCAGCAGCAGCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................AGCAAC------A-------AC----AACAACAG---CA-----GC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................AGCAGCAACAACAACAGCAAC---------...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GCAAC------A-------AC----AACAACAG---CA-----GCA---------G------CAGT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CAGCAAC---------AACAGCAGCAGC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CA-----GCA---------G------CAGCAGCAACAACAA------C-----------A............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CAGCAGCAACAACAA------C-----------AACAG........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................AC------A-------AC----AACAACAG---CA-----GCA---------G------CAGCAGC....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CAGCAAC---------AACAGCAGCAGCAAC------A-------......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAACAGCAAC---------AACAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................GCAAC---------AACAGCAGCAGCAAC------A-------A........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |