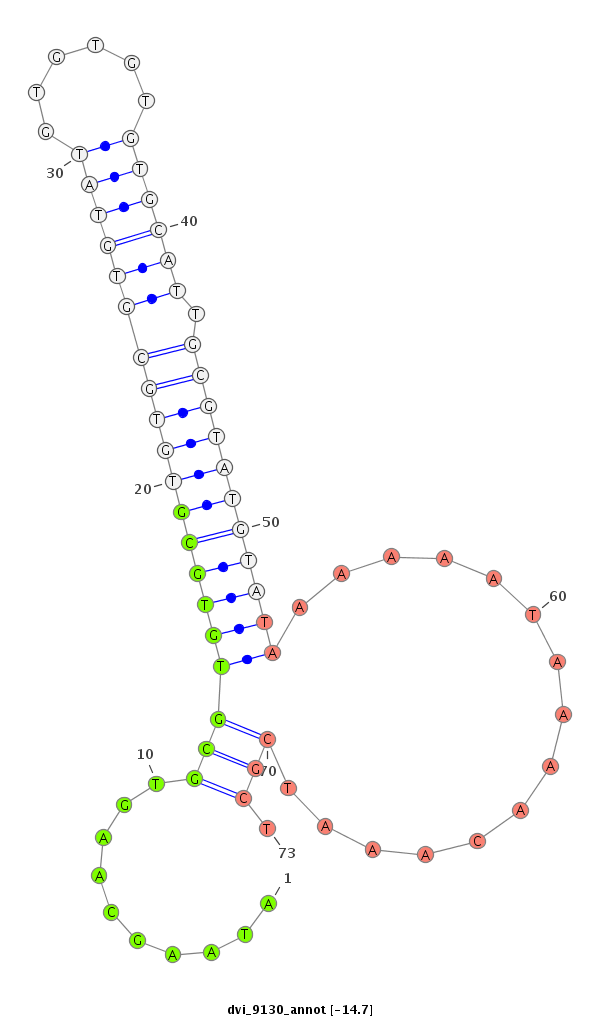
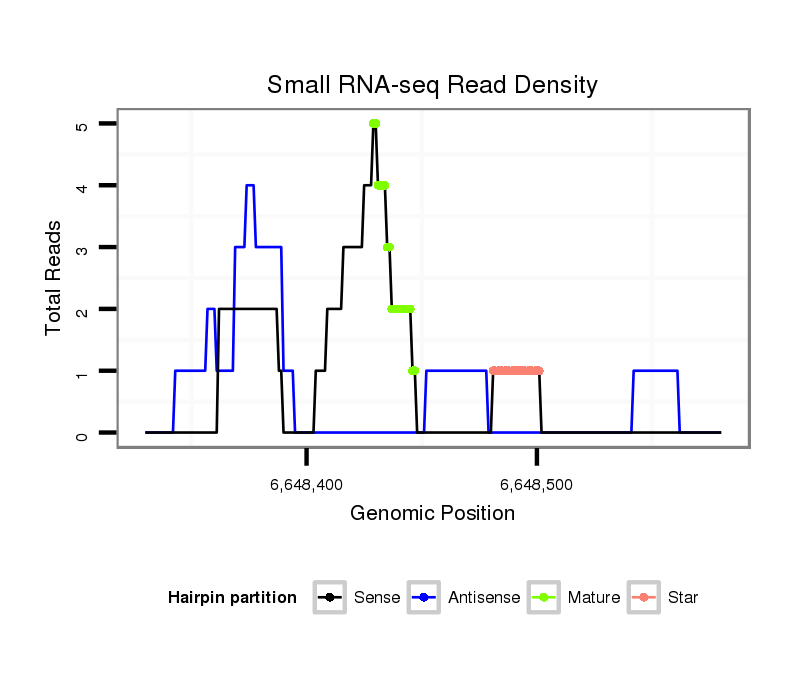
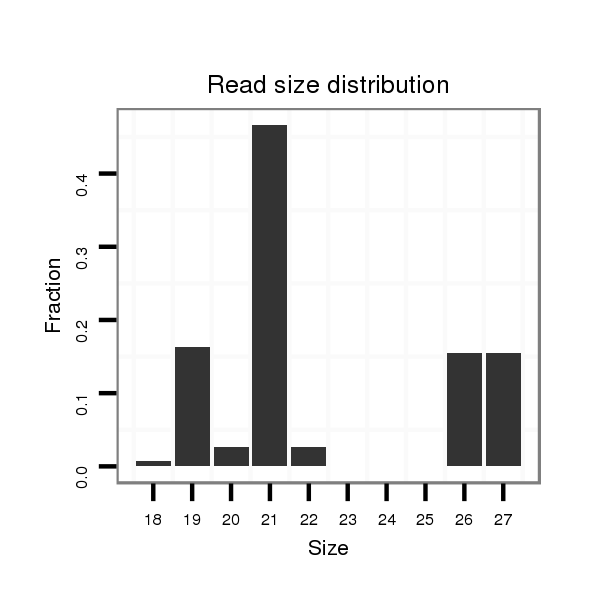
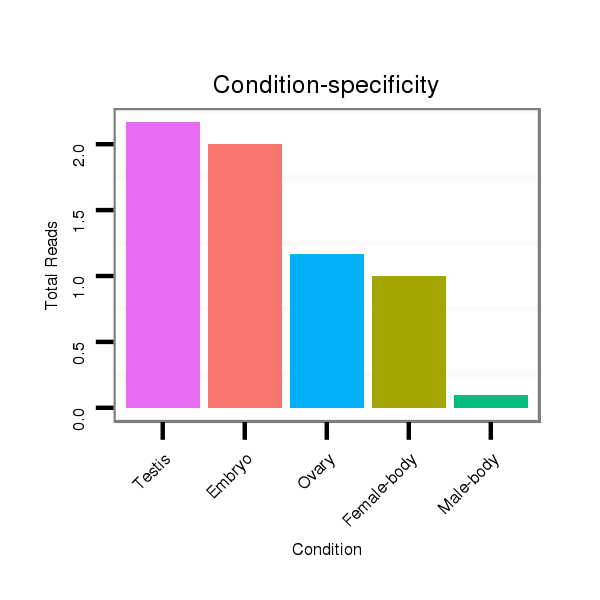
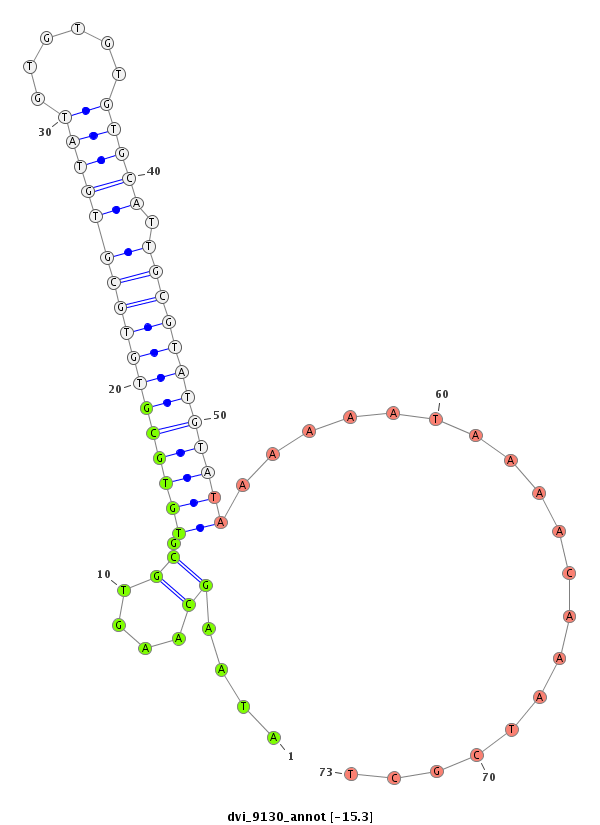
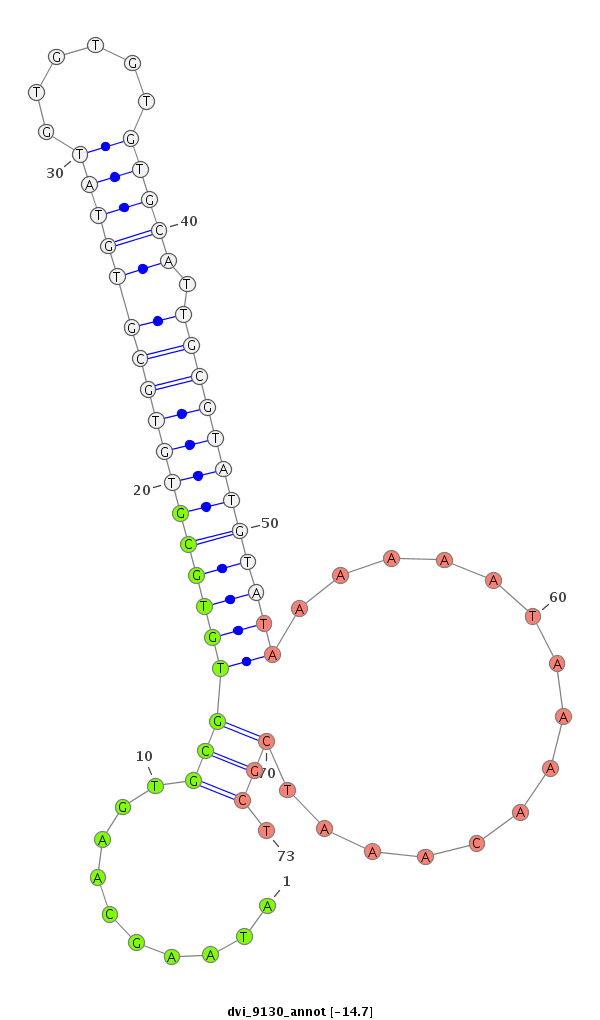
ID:dvi_9130 |
Coordinate:scaffold_12928:6648380-6648530 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.3 | -15.3 | -14.7 |
 |
 |
 |
CDS [Dvir\GJ17022-cds]; exon [dvir_GLEANR_17527:1]; intron [Dvir\GJ17022-in]
| Name | Class | Family | Strand |
| (TG)n | Simple_repeat | Simple_repeat | + |
| ------------------------------####################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTAAGCTTACACCAGCCCTTCTAAACCAACATGTCGTTTTTTCGCTATGTGTAAGAGAAGGTCAGAAAAATGTACATACATATGTGTAGTGGAAGGTGCATAAGCAAGTGCGTGTGCGTGTGCGTGTATGTGTGTGTGCATTGCGTATGTATAAAAAATAAAACAAATCGCTAAGCGATTAACAACAAATTAAAAGTGCAAAGTAATCACTGTTAGTACATATATGAGCTTATATGTGTATGTACATGTAT ***************************************************************************************************..........((((((((((((((((((((......)))))).)))))))))))...............))).******************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060674 9x140_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
V053 head |
SRR060678 9x140_testes_total |
M028 head |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060687 9_0-2h_embryos_total |
M061 embryo |
SRR060679 140x9_testes_total |
SRR060688 160_ovaries_total |
GSM1528803 follicle cells |
V116 male body |
SRR060689 160x9_testes_total |
SRR060666 160_males_carcasses_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GTCGTTTTTTCGCTATGTGTAAGAGA................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ATAAGCAAGTGCGTGTGCG..................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................GTCGTTTTTTCGCTATGTGTAAGAGAAG............................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CGGAAGGTGCAGAAGTAAGTGC............................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................GTGTGTAGTGGAAGGTGCATA..................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GTGCATAAGCAAGTGCGTGTG....................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CGCTATGTGTAAGAGAAGGTCC........................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TAAAAAATAAAACAAATCGCT............................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CATACATATGTGTAGTGGAAGGTGCAT...................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................ATATGTGTAGTGGAAGGTGCATAAGC.................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CTTTTTTTCGCTATGTGTAAGAGAA................................................................................................................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TAGTGGAAGGTGCATAAGCAA................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TGTGTGTGCGTTGCGTATGTA.................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GGGCGTGTGCGTGTATGTGTGTGTTC................................................................................................................ | 26 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TGAGACGGTCAGGAAAATGT.................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GCGTGTGCGTGTGCGTGTAT.......................................................................................................................... | 20 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TGTGCGTGTATGTGTGTGTGCA............................................................................................................... | 22 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GTGCGTGTATTTGTGTGTGCA............................................................................................................... | 21 | 1 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CAAATGGCTAAGCGGTTAAAA................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................TGAAGGTGAGAAAAATGTAGA............................................................................................................................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................GAAGGTGAGAAAAATGTAGAG.............................................................................................................................................................................. | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GTGCGTGTATGTGTGTGTCCA............................................................................................................... | 21 | 1 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........ACCAGCCCTCCTTCACCAA.............................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................GTGTGCGTGTGCGTGTAT.......................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................GCATGTGTGTGTGCATTG............................................................................................................ | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................GTGCGTGTGCGTGTGCGTG............................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CATTCGAATGTGGTCGGGAAGATTTGGTTGTACAGCAAAAAAGCGATACACATTCTCTTCCAGTCTTTTTACATGTATGTATACACATCACCTTCCACGTATTCGTTCACGCACACGCACACGCACATACACACACACGTAACGCATACATATTTTTTATTTTGTTTAGCGATTCGCTAATTGTTGTTTAATTTTCACGTTTCATTAGTGACAATCATGTATATACTCGAATATACACATACATGTACATA
*******************************************************************************..........((((((((((((((((((((......)))))).)))))))))))...............))).*************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060673 9_ovaries_total |
M047 female body |
SRR060675 140x9_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
SRR060689 160x9_testes_total |
SRR060661 160x9_0-2h_embryos_total |
V116 male body |
SRR060683 160_testes_total |
SRR060654 160x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................AAAGCGATACACATTCTCTTC............................................................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................GATACACATTCTCTTCCAGTC.......................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............TCGGGAAGATTTGGTTGT............................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GCACATACACACACACGTAACGCATAC...................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TTGTACAGCAAAAAAGCGATA........................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AATCATGTATATACTCGAAT................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GTTTACCGATTCGCCAATT..................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............GTTGGGAAGGTTTAGTTGTAC.......................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....CGAATGCGGTTGTGAAGAT.................................................................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................TGTTGTTAAATTTGCAGGTT.................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:6648330-6648580 + | dvi_9130 | GTAAGCT---TACA-----CCAGCCCTTCT-----------------A---------AACCAACATGTC--GT---TTT-TTCGCT------------------------ATGTGT-AAGAGAAGGTCAG------AAAAAT-----------GTACAT--------AC----AT-A--------------------------------------TGTGTAGTGGAAG-------------GTGCATAAGCAAGT----GCGTG-TG-CGTGTGCGT--------------------------------------------------------GTATG----TG----TGTGT----------GCA----------------------------------------------T---------------TG-----------CGT------------ATGTAT------------AAAAAATAAAACAAATCGC------------TAAGCGATTAACAAC----------AA---------------------------ATTAAAAGTGCAAAGTAATCACTGTTAGTACATATATGA----GCTTATATGTGTATGTACATGTAT |
| droMoj3 | scaffold_6328:2331169-2331426 + | GTGTGCT---TACG-----CTGGTCCGTCT-----------------A---------CAGCAATATATGTATT---TTT-TTCTCT------------------------ATGTGC-AAGAGAAGGTCAG------AAAAAT-----------GTACAC--------ACATACAT-A--------------------------------------TGTGTAGTGGAAG-------------GTGCATAACCAAGT----GCGTG-TG-CGTGTGTGT--------------------------------------------------------ATGCG----AGTGTTTGTGT----------GCA----------------------------------------------T---------------TG-----------CGT------------ATGTAT---------CAG--AAGAAGAAACAATTCGC------------TAAGCGTTTAACAAC----------AA---------------------------ATTAAAAGTGCAAAGTAAGCACT----GTTTGTACATAT----ATGAGCTCATTTGCGTATGTGTAC | |
| droGri2 | scaffold_14853:7221937-7222187 - | GTGAACT---AACA-----A--AACCGTGC-----------------A---------ACGCAAAATGTCTATT---TTC-CTCACC------------------------GTGTGT-AAGAGAAGGTCAG------AAAAAT-----------GTACAT--------TC----AT-A-----------------------TG-------------TGTGTAGTAGAAG-------------GTGCATAAGCAAGT----GCGTG-TG-AGTGTATCT------------------------------------------------------------G--------TATGTGT----------GCA----------------------------------------------T---------------TG-----------CGT------------ATGTAT------------AAAAA--AAAACAAATCGC------------AATGCGATTAACAAC----------AA---------------------------AATAAAAGTGCAAAGTAAGCACTGTTAGTACATACATAAACATACACATATCTGTGTGTATATGCGC | |
| droWil2 | scf2_1100000004963:2406765-2407067 - | ACAAGCA---TACA-----CCA---TGTGC-----------------AACTTATTAATATTAACT--TC--AC---CTA-CACATC------------------------AAGTGT-AAGAGAAGATCCGAGTCAAATCACT-----------GTGCGAGGAATTTAAG----TT-C--------------------------------------TGTG-AGTA-----------------GTGTGTG-----------------CG-T----C------TATAGG----------------TGTATATGTCGTGTC--GT--------------------------------GTA--------TG------------------------------TGTGTGTGTGTGAGTGTAGGGCTAATA-CAGCTG-----------CGT------------ATGTATTACC-------G--AAAA--TAACAA------------------ATGTGTTCGACAGTAGCAACAACAAAAATTCAATTACCAAACGAGAATCACTACGAAAAACTGCATAGTAA-----------------GCAT----CTGTATATGTATGTTTACAAAAAT | |
| dp5 | XL_group1e:293926-294178 - | GGCAATT---TGC-AATTT-----------------------------------CAGTGTATAAATGCC--GTTTT---ACTTGTA------------------------ATGTTCAAAGAGAAGGTCAG------AAAAAT-----------GTACAC--------AG----TT-C--------------------------------------TGTGTAGTAGCAGCA-----------GTGCATA------------------GAT-----------TGTAGG----------------CATACCTGCGCTCCCAAGT--------------------------------GTA--------AG------------------------------TGTGTGCGTGTGCGCATAATGTT-ATA-CAGCTGTCGCCGCCTATCGT------------GTGTAT---------CAG--TGAA--TAACAA------------------ATGTGATCAACAAA----------AA---------------------------------AGTGCATTGTAAGCATTGTT----------------------AATGCATTCACATACACG- | |
| droPer2 | scaffold_60:37291-37543 + | GGCAATT---TGC-AATTT-----------------------------------CAGTGTATAAATGCC--GTTTT---ACTTGTA------------------------ATGTTCAAAGAGAAGGTCAG------AAAAAT-----------GTACAC--------AG----TT-C--------------------------------------TGTGTAGTAGCAGCA-----------GTGCATA------------------GAT-----------TGTAGG----------------CATACCTGCGCTCCCAAGT--------------------------------GTA--------AG------------------------------TGTGTGCGTGTGCGCATAATGTT-ATA-CAGCTGTCGCCGCCTATCGT------------GTGTAT---------CAG--TGAA--TAACAA------------------ATGTGATAAACAAA----------AA---------------------------------AGTGCATTGTAAGCATTGTT----------------------AATGCATTCACATACACG- | |
| droAna3 | scaffold_13117:5608247-5608480 - | GTGAATG---ACC--------A---TTTTA-----------------AAGTTGT-------------TC--CTTTT---GGTTCAA------------------------AACGTCGCCGAGAAGGTCAGAATCAGAAAAAT-----------GTACAC--------AG----TTTG--------------------------------------TGTGT---------------------------------GCCAGTGTGTG-TG-TGTGTGTGT--------------------------------------------------------GTGTG----AG----TGAGTGTATGTGTGTGTATGATATAGGGAAGAGGGGGGTAGGAGTACAATTGTGTGAGTGCGCAGAAAGTT-ATA-CAGCTG----------GCGT------------ATGTATCAGCGAAATCAG--TAAA--TAACAA------------------CTGTGAT----------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396540:42920-43092 + | ATGAGAT---AACG-----T--------------------------------------------------------TAC-TTTAC------------------------------------------------------TAT-----------ATGTAT--------AT-------------------------------------------------------------GTATATGTATA-----------TGT----ATAT-GT---------ATATGTATATGTATATGTATATGTATATGTATATGC------ATATGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------G-----------TATATGTATATGTATATGT----------------------------------------------------------------------------------------------------------------------------ATATGTATATGTAT----ATGTATATGTATATGTATATGTAT | |
| droKik1 | scf7180000302469:1786711-1786933 - | AACTTATCCTCGC-ACTTG-----------------------------------CGAAATAAGCATTCC--GAAAT---TTTCGCA------------------------ACATG--------CGGTCAA------AAA-AG-----------GTATGT--------ACATATGT-A--------------------------------------TGTAT-------------------------------GTTG----GCATA-TG-TGTGTATGTG--------------------------------------------------------GATG----TGCG------------------------------------------------------------------TTGAGG----------TG-----------CAT------------ATGCAT------------GCATA--AATGCAAATTAAATCGAAATTTAACACGTGGTTTACTTC----------TA---------------------------------ATTTCATTTTT--------AGCTGTATGTATGT----TGGCATATGTGTATGTATGTATGT | |
| droFic1 | scf7180000453901:452436-452604 - | GA-----------------------------------------------------------------------------------------------------------------------GAAGGTCAG------AAAAAT-----------GTACAC--------AG------------------------------------------------------------------------GCGCATATCTGCGG----CCGTG-TG-TGTGTTCGTG--------------------------------------------------------------------------------------CG------------------------------TGCGTGTGAGTGCGCAGAAAGTT-ATA-CAGCTGTCATCGACTGGCTT------------ATTTAT---------CAA--TAAA--TAACCA------------------CTGTAACC----------------------------------------------------------------------GTCAGAAAACGCCA----GTTACTGGGTAAATGCGTATATAA | |
| droEle1 | scf7180000491006:2881512-2881693 + | GA-----------------------------------------------------------------------------------------------------------------------GAAGGTCAG------AAAAAT-----------GTACAC--------AG------------------------------------------------------------------------GCGCATACCTGCGC----CCGTG-TG-TGTGTGTGTG--------------------------------------------------------TGTG----TCCGTATGTG--AACG------TG------------------------------TGCGTGTGAGTGCGCAGAAAGTT-ATACCAGCTGTCATCGACTGGCGT------------ATTTAT---------CAA--TAAA--TAACTA------------------CTGTGATCAACAGA----------AA----------------------------------------------------------------------ACTTCAGTTACTGGGTAATTGCAT | |
| droRho1 | scf7180000770833:655-847 - | CATAT-----------------------------------------------------------------------------------------------------------------------ATATGT------ATATAT-----------GTATAT--------AT--------GTATATATGTATATATGTATATATGTAT---------------------------ATATGTATA-----------TAT----GTAT-AT---------ATGTATATATGTATATATGTATATATGTATATATGT------ATATATGTATATATGTATATATGTATAT------------------------------------------------------------------------------------A-----------TGTATATATGTATATATGT----------------------------------------------------------------------------------------------------------------------------ATATATGTATATAT----GTATATATGTATATATGTATATAT | |
| droBia1 | scf7180000300828:85-257 - | GCTGTTTCTTCGT----T--------------------------------------------------------TT---TGCTG-------------------------------------AAAACTTAG------AAAAATAGACGGAGCGTATATGT--------ATATATAT-A--------------------------------------TGT-----------------------------------------------------------------------ATGTATATGTATATGTATATGT------ATATGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------G-----------TATATGTATATGTATATGT----------------------------------------------------------------------------------------------------------------------------ATATGTATATGTAT----ATGTATATGTATATGTATATGTAT | |
| droTak1 | scf7180000415864:222939-223032 - | A---------------------------------------------------------------------------------------------------------------------------GGTCAG------AAAAAT-----------GTACAC--------AG------------------------------------------------------------------------GCGCATACCTGCGC----CCGTG-TG-TGTGTGCCT--------------------------------------------------------GTGCC----TG----TGT--GTG--------TG------------------------------TGCGTGTGAGTGCGCAGAAAGTT-ATA-CA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409801:544809-545026 + | ATATCTC---TG-------CT---------GGTTTAGTAGCAGTATCA---------AATTAAAAATTC--GA---TTT-TTTCTTGCACGAAATAAGCTTGGATCCAC-----------------------------------------------------------T-------------------------------------------------------------GTATATATATA-----------TAT----ATAT-AT---------ATATATATATGAATATGTATATTTCTATGTATGTGT------ATATGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------G-----------TGTATGTAAGTGTATATGC----------------------------------------------------------------------------------------------------------------------------AAGTGTATATGTAA----GTGTATATGTAAGTGTATATGTAT | |
| dm3 | chrUextra:26950111-26950358 - | AACAACT-----------------------GCAACAGCACCAACACCA---------ACT-------------------CT-------------------ATGCTCCACTATATGT-AT-----ATATGT------ATATAT-----------GTATAT--------AT--------GTATATATGTATATATGTATATATGTATATATGTATAT---------------ATGTATATATG-----------TAT----ATAT-GT---------ATATATGTATATATGTATATATGTATATATGTATAT------ATGTATATATGTATATATGTATATATGT------------------------------------------------------------------------------------A-----------TATATGTATATATGTATAT----------------------------------------------------------------------------------------------------------------------------ATGTATATATGTAT----ATATGTATATATGTATATATGTAT | |
| droSim2 | x:3698005-3698166 + | GA-----------------------------------------------------------------------------------------------------------------------GAAGGTCAG------AGAAAT-----------GTACAC--------AG------------------------------------------------------------------------GCGTAAATTTGTGT----GCGTG-TG-TGTGGGTGCG--------------------------------------------------------TACA----TC----CGT--GT--------GCG------------------------------CTCGTGTGAGTGCGCAGAAAGTT-ATA-CAGCTGTCATCGACTGGCGT------------ATTTAT---------CAA--TAAA--TGACTA------------------CTGTGATCAGCGAA----------AA-----------------------------------------------------------------------------------ACTTCAGTTAC | |
| droSec2 | scaffold_12497:147-313 - | GACAA-----------------------------------------------------------------------------------------------------------------------ATGTAT------ATGTAT-----------ATGTAT--------AT-------------------------------------------------------------GTATATGTATA-----------TGT----ATAT-GT---------ATATGTATATGTATATGTATATGTATATGTATATGT------ATATGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------G-----------TATATGTATATGTATATGT----------------------------------------------------------------------------------------------------------------------------ATATGTATATGTAT----ATGTATATGTATATGTATATGTAT | |
| droYak3 | 4:399999-400162 + | ACGAAAC---CGTA-----TAAATCCACGT-----------------G---------TTATGAAATGCGTG-----------------------------------------------------------------------------------------------------------------------------AATAAATCT----------------------------------------------TAAA----TCTAA-A-----------------------ACGAATGCATATATGTATATGT------ATATGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------G-----------TATATGTATATGTATATGT----------------------------------------------------------------------------------------------------------------------------ATATGTATATGTAT----ATGTATATGTATATGTATATGTAT | |
| droEre2 | scaffold_4929:25044455-25044640 + | TATGTGT---AGTG-----T------------------------------------------------------------------------------------------------------ATGTACTG------GAGTAT-----------GTATGT--------AT----GT-A-------------------TGTATGTAT----------------------------------------------GTAA----TCATG-TA-TGTATGTATGTATGTATGTATGTAGGCATGTATGTATGTATGT------ATGCTTGTACACATGTATGTATGTATGT------------------------------------------------------------------------------------G-----------TGTATGTATTGGTGTATGT----------------------------------------------------------------------------------------------------------------------------ATGTATGTATGTAT----GTGTGTATGTATTGGTGTATGTAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/19/2015 at 03:28 PM