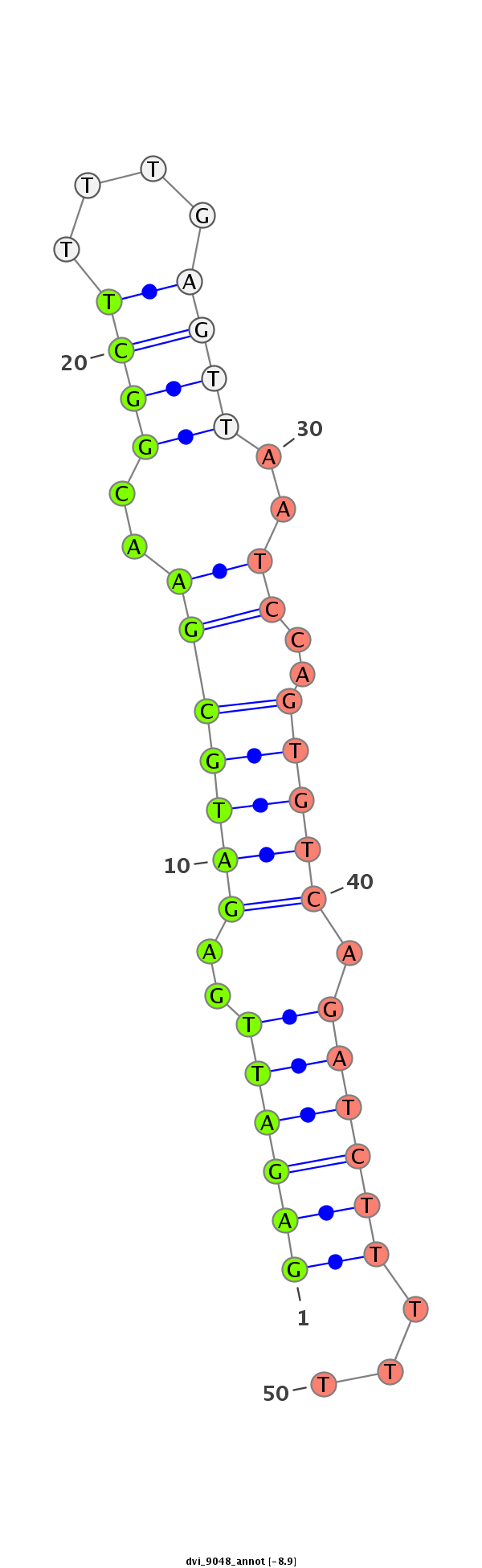
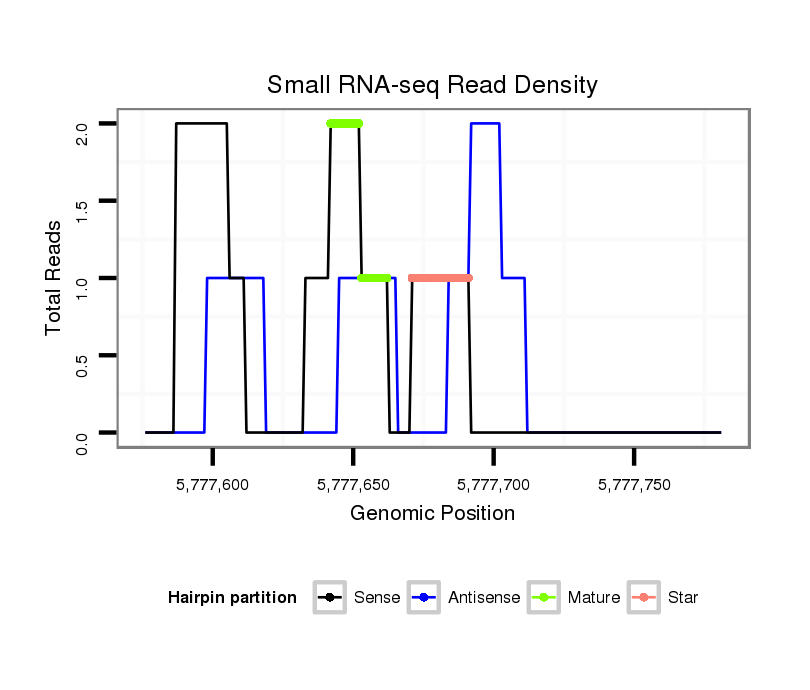
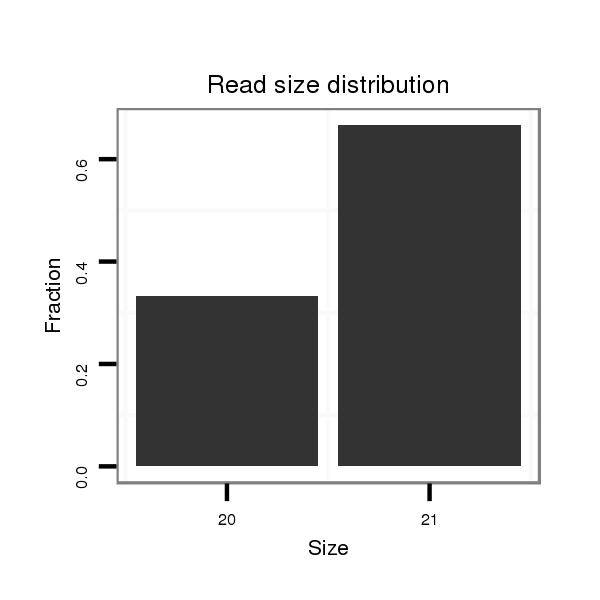
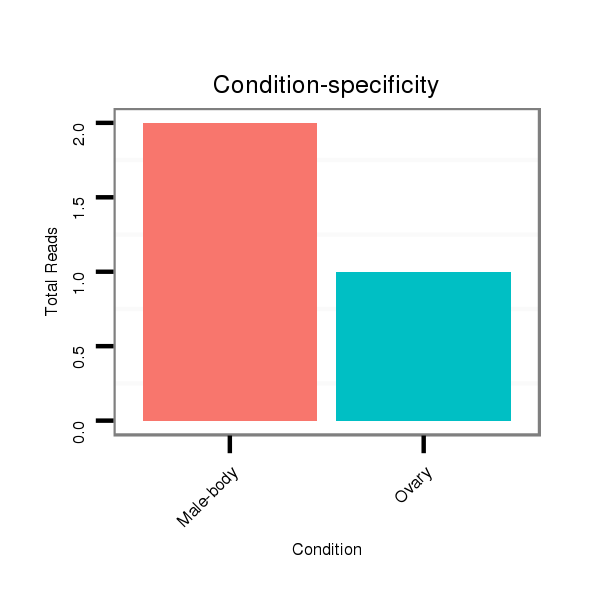
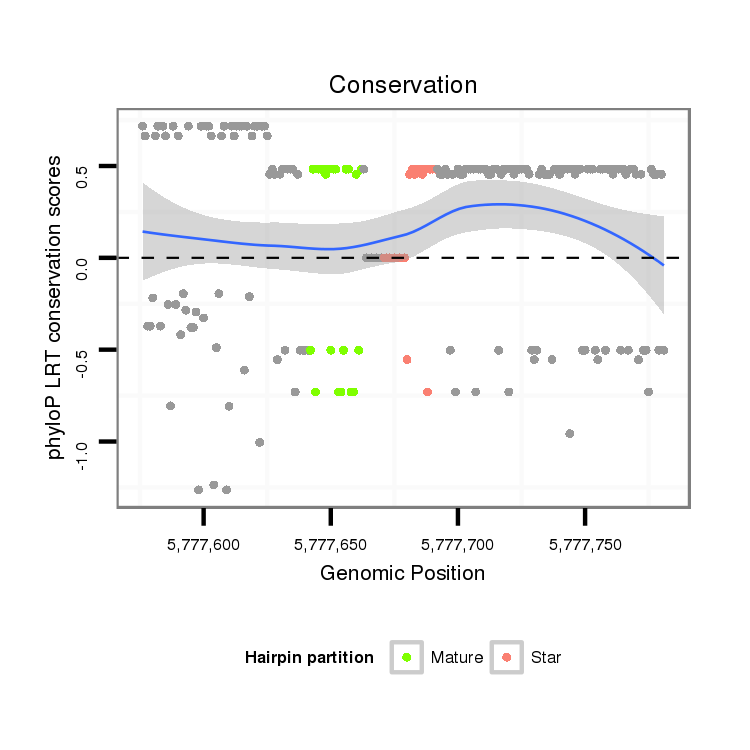
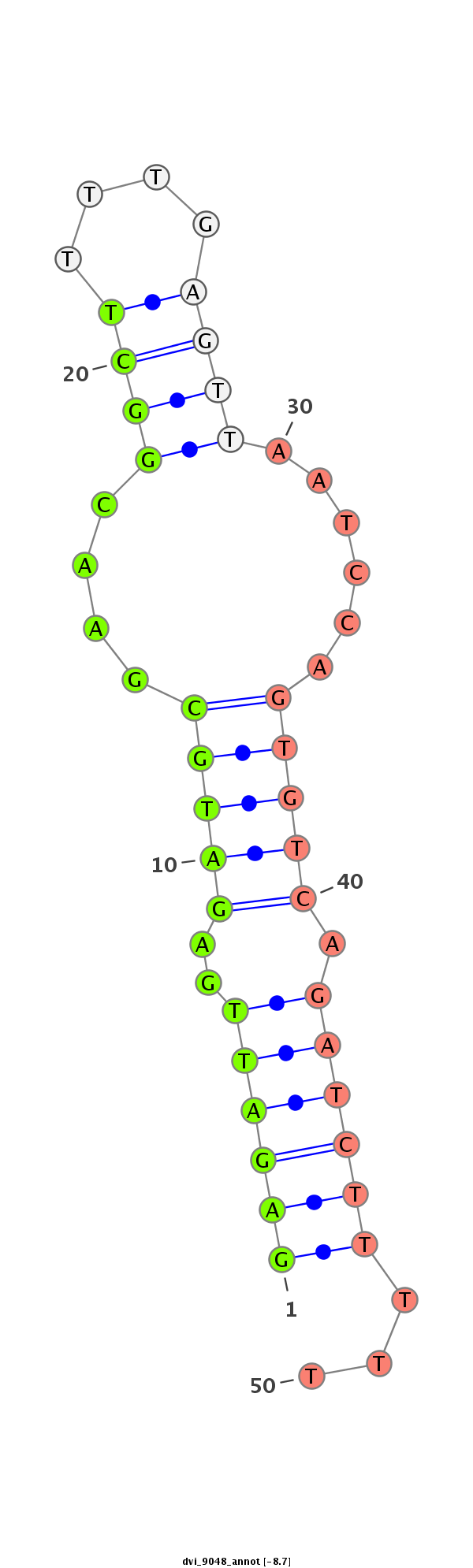
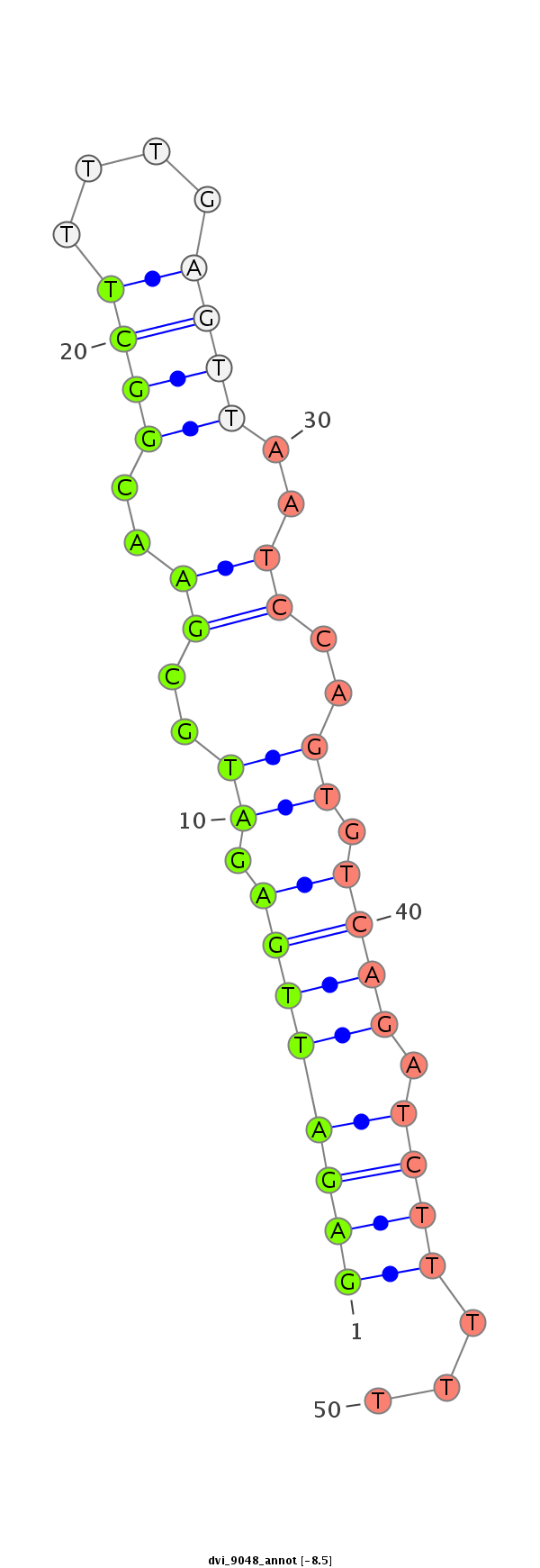
ID:dvi_9048 |
Coordinate:scaffold_12928:5777626-5777731 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.7 | -8.5 | -8.5 |
 |
 |
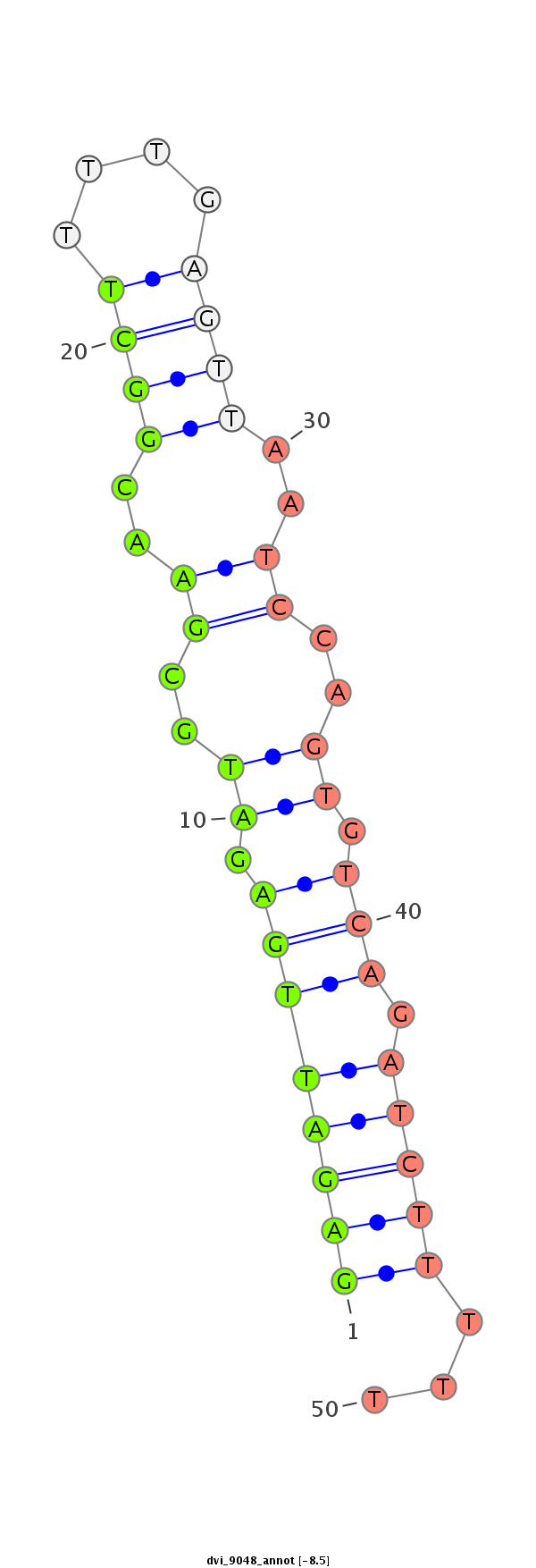 |
CDS [Dvir\GJ16977-cds]; exon [dvir_GLEANR_17485:2]; CDS [Dvir\GJ16977-cds]; exon [dvir_GLEANR_17485:1]; intron [Dvir\GJ16977-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------------------------------------------################################################## ACGGGGACTGTCTTCTGATAACGACAACCGCGTGGAGATTCAAGAATTTGGTGTGTAAATCGCACCGAGATTGAGATGCGAACGGCTTTTGAGTTAATCCAGTGTCAGATCTTTTTAGCTGCCAAGCAAATCTATTGCTATGAAATGATATTGCAGGAGCGAAAGATTGTGAATGATAGTTTCATCAAGTGCATCACAACTGCCGA ******************************************************************((((((..(((((((..((((....))))..))..))))).))))))...****************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060686 Argx9_0-2h_embryos_total |
GSM1528803 follicle cells |
M027 male body |
SRR060656 9x160_ovaries_total |
SRR060657 140_testes_total |
SRR060674 9x140_ovaries_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................GATTGCAGGAGCGAAATATCGTG................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CATGAAATGAGCTTGCAGGAGC.............................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........CTTCTGATAACGACAACCGCGTGGA.......................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TGCAGGACCGAAAGATTGTGAAT................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................GAGATTGAGATGCGAACGGCT....................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TCAGATCTTTTTGGCTGCCAGG................................................................................ | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GACCGAAAGATTGTGAATG............................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........CTTCTGATAACGACAACCG................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AATCGCACCGAGATTGAGAT................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TTGCAGGAGCGAAGGATTGTG................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................AATCCAGTGTCAGATCTTTTT.......................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GATTGCAGGAGCGAAATAT....................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GATTGCAGGAGCGAAATATC...................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ACAGGGTCTTTCTTCTGAT........................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................GCAAATCTATCGCTTTGA............................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TGCCCCTGACAGAAGACTATTGCTGTTGGCGCACCTCTAAGTTCTTAAACCACACATTTAGCGTGGCTCTAACTCTACGCTTGCCGAAAACTCAATTAGGTCACAGTCTAGAAAAATCGACGGTTCGTTTAGATAACGATACTTTACTATAACGTCCTCGCTTTCTAACACTTACTATCAAAGTAGTTCACGTAGTGTTGACGGCT
******************************************************************************************((((((..(((((((..((((....))))..))..))))).))))))...****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M047 female body |
SRR060654 160x9_ovaries_total |
SRR060664 9_males_carcasses_total |
GSM1528803 follicle cells |
SRR060662 9x160_0-2h_embryos_total |
V053 head |
SRR060678 9x140_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................CTGTTGGCGCACCTCTAAGTT................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TAGAAAAATCGACGGTTCG............................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TCGACGGTTCGTTTAGATAA...................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................TAACTCTACGCTTGCCGAAAA.................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGGCTCTAACACTACGCTAT........................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TAGTTTACGTAGTGATGAC.... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................CGATCCTTTACTATAA...................................................... | 16 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................ACTTAATTAGGTCACAGACA................................................................................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................ATTAACGATACTTTACTA......................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:5777576-5777781 + | dvi_9048 | ACGGGGACTGTCTTCTGATAACGACAACCGCGTGGAGATTCAAGAATTTGGTGTGTAAA-----------------TCGCACCGAGATTGAGATGCGAACGGCTTTTGAGTTAATCCAGTGTCAGATCTTTTTAGCTGCCAAGCAAATCTATTGCTATGAAATGATATTGCAGGAGCGAAAGATTGTGAATGATAGTTTCATCAAGTGCATCACAACTGCCGA |
| droMoj3 | scaffold_6359:4284144-4284350 + | ACAAAGATTGCGTCCAGGTAACCAAAACGGCGTCCAGATTGAAGAAGTTGGTGAGTGAAGTCATCAGATCATCAGTTAGTGTTAATATTGAAATTAAAAATGTTT----------------ACAGATCTGTTTAGCTGTCCAGCAAATATATTGCTACGAACTGATATTGTTAGAGCGTAAGATTCTGAACAATAAATTTATCAAATGTATCTCGGATGCTGG | |
| droGri2 | scaffold_14853:4910348-4910397 + | ACAAGGATTGTGTTCAAGTCCATACAACTCTGTTCAGATTCAGGAAATTG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 03:18 PM