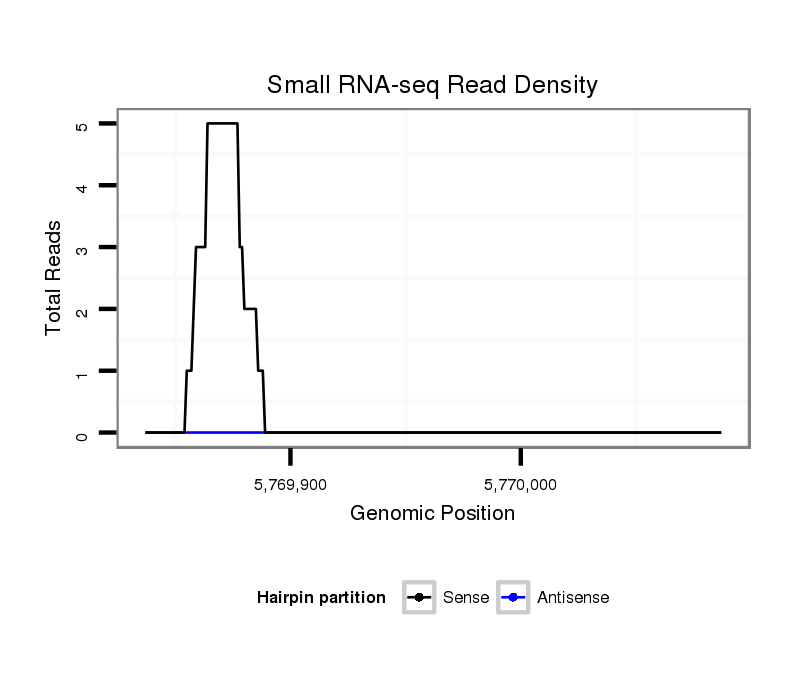
ID:dvi_9046 |
Coordinate:scaffold_12928:5769887-5770037 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_17484:1]; CDS [Dvir\GJ16976-cds]; intron [Dvir\GJ16976-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGAATACAAAACAACTGGCAAGGTAACGCCCAATCCGGACCAGAACTCAGGTAATAGCCGATCAACTTATTTGAAAGAAACCTCAGCTTAAGCTTTATCTATGCTTAGGTCTTATCATTTTCAATTCCATATCTAATCATGTTCATTTTGCCAACAAAAAAAAAAAGCTTTCTTTAAGATTCTTGTAATGTTGTCAAGATTATTTTTGATAGAAAGCTTATAGAGGATTCGATGTACAGCGAAATTTTGAA **************************************************....................(((((((........((((((((.........)))))))).......)))))))....................((((((.(((((...........(((.....)))..........))))).)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
SRR060679 140x9_testes_total |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
V116 male body |
SRR060674 9x140_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................AAAAAAAAAAGCTCACTGTAAGAT....................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................AAAAAAAAAAAGCTCACTGTAAGAT....................................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CAAGGTAACGCCCAATCCGGACCAG................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GTAACGCCCAATCCGGACC.................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................GCCCAATCCGGACCAGAACTCA.......................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................GCCCAATCCGGACCAGAACTCAGGT....................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................GGTAACGCCCAATCCGGACC.................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TAAAAAGCTTTCTTTGAGA........................................................................ | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ACAAAATAACAAAGCTGTCTTT............................................................................ | 22 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................TCAGTTTGGCAACATAAAAAA........................................................................................ | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................AGGAGTGGAAGTACAGCGA......... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................TATTATTCCAACAAAAAAAAAAAG.................................................................................... | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CCTTATGTTTTGTTGACCGTTCCATTGCGGGTTAGGCCTGGTCTTGAGTCCATTATCGGCTAGTTGAATAAACTTTCTTTGGAGTCGAATTCGAAATAGATACGAATCCAGAATAGTAAAAGTTAAGGTATAGATTAGTACAAGTAAAACGGTTGTTTTTTTTTTTCGAAAGAAATTCTAAGAACATTACAACAGTTCTAATAAAAACTATCTTTCGAATATCTCCTAAGCTACATGTCGCTTTAAAACTT
**************************************************....................(((((((........((((((((.........)))))))).......)))))))....................((((((.(((((...........(((.....)))..........))))).)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1106729 mixed whole adult body |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
SRR060660 Argentina_ovaries_total |
SRR060667 160_females_carcasses_total |
M027 male body |
SRR1106728 larvae |
SRR060663 160_0-2h_embryos_total |
SRR1106727 larvae |
V053 head |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................CATAGATTAGGACAAG........................................................................................................... | 16 | 2 | 20 | 1.30 | 26 | 23 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 |
| ..............................................AGTCCATTAACGTCTAGTTGA........................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ATAGATTAGGACAAG........................................................................................................... | 15 | 1 | 13 | 0.23 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ATAGATTAGGACAAGTGA........................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTCTGGTCGAAAGAAATTCTA....................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TCAAGAGTAGTAAAAGTTCAG............................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CGGGGTAGGCCTGATCGTG............................................................................................................................................................................................................. | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................TCGCGGGTTAGGCGTGATC................................................................................................................................................................................................................ | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................CGCTCCCTTGCGGGCTAG........................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................ATAGATTAGGACAAGCAAT....................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................AGGTATACATTAGTACCAC........................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................AAGGACATTATAACCGTTC..................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:5769837-5770087 + | dvi_9046 | GGAATACAAAACAACTGGCAAGGTAACGCCCAATCCGGACCAGAACTCAGGTAATAGCCGATCAACTTATTTGAAAGAAACCTCAGCTTAAGCTTTATCTATGCTTAGGTCTTATCATTTTCAATTCCATATCTAATCATGTTCATTTTGCCAACAAAAAAAAAAAGCTTTCTTTAAGATTCTTGTAATGTTGTCAAGATTATTTTTGATAGAAAGCTTATAGAGGATTCGATGTACAGCGAAATTTTGAA |
| droMoj3 | scaffold_6359:4275708-4275763 + | GGAATACAAAACAACTGGCAAAGTTACGCCCAATCCGGATCAGAATTCAGGTATTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14853:4902686-4902760 + | GGAATACAAAACGACTGGCAAAATAACGCCCAATCCGGAACAGAACTCAGGTAATACGCCAAAAACTGATTTGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:12219871-12219923 - | ATGC----------------------------------------------------------------------------------------------------TCGGGTCCAATCACTTTGTATTCCATATTAATTTCTGTTGCTGCTGCTA-------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/19/2015 at 03:18 PM