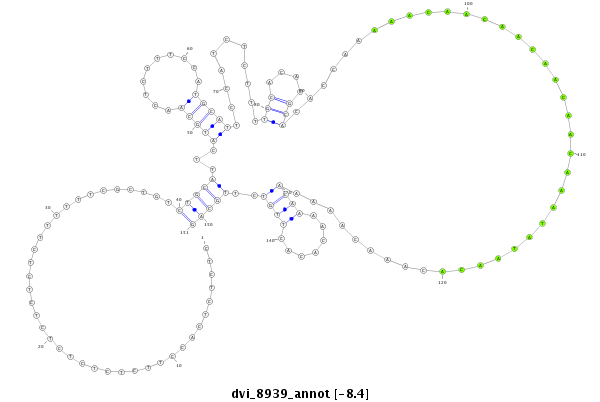
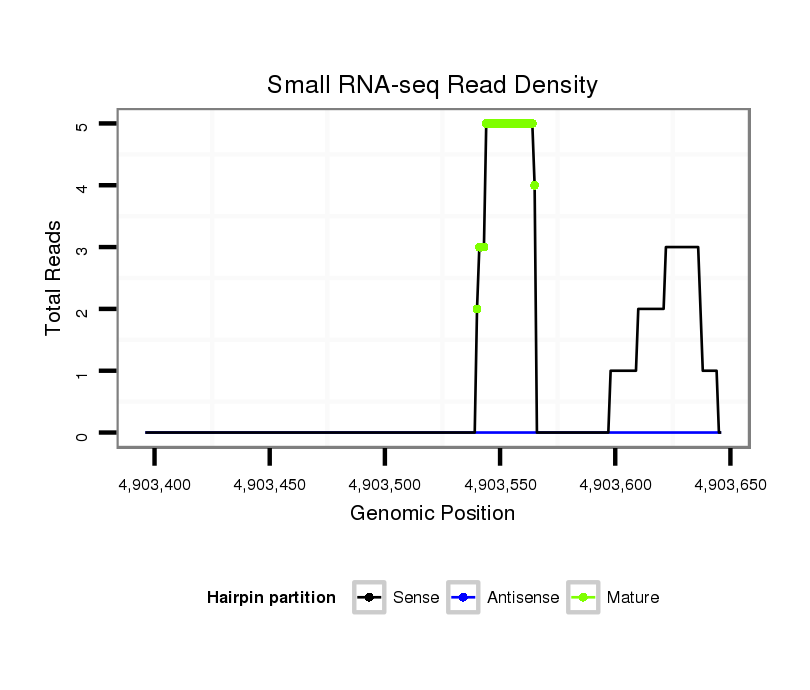
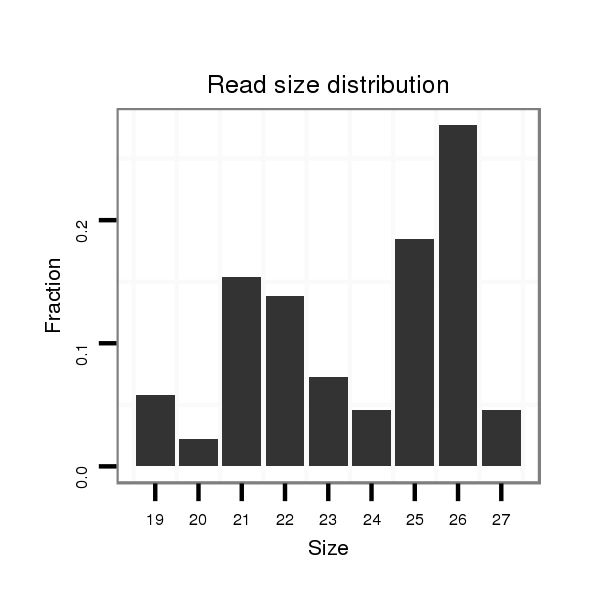
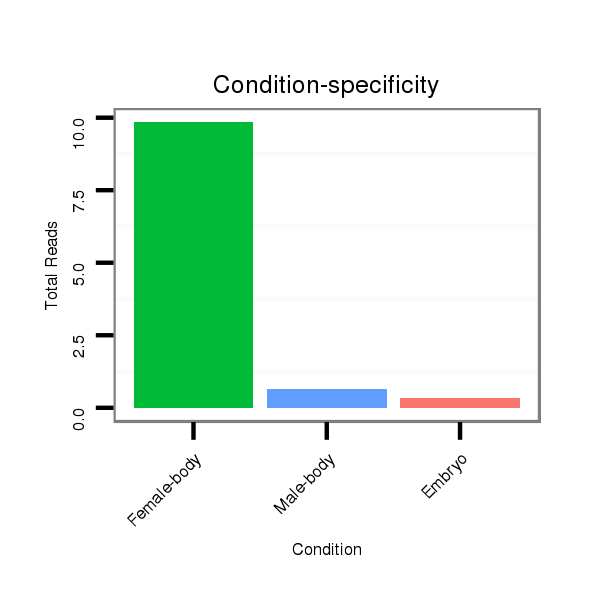
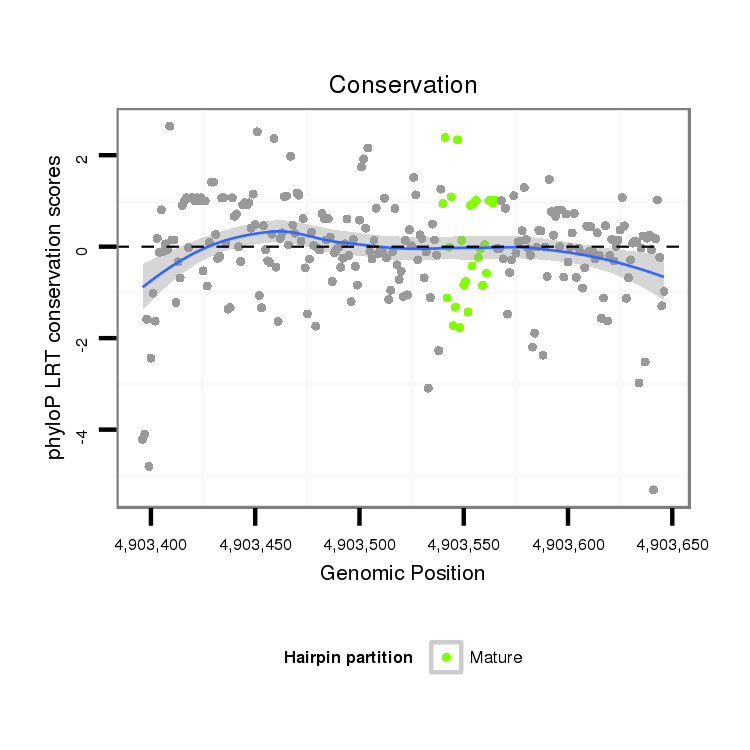
ID:dvi_8939 |
Coordinate:scaffold_12928:4903446-4903596 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.3 | -7.7 | -7.6 |
 |
 |
 |
CDS [Dvir\GJ16942-cds]; exon [dvir_GLEANR_17447:4]; intron [Dvir\GJ16942-in]
| Name | Class | Family | Strand |
| (TC)n | Simple_repeat | Simple_repeat | + |
| (CAAAAA)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GAAACAATTTATTTGCTCTACTAATCACATGTAATATATTCCATCTCTCTCTCTCTCACCTTCTCTCTCTCTCTCTCTTTTTTCGCTGTCTGCATTCATGCAACTCTTTCCATGCATTCCATCTCTTTTGCACACGCACACCAAAAACAACAACAACAACAAATATAACACAAACAAAAACAAAACACACTTGTCTTGCAGTGAGCTATGCGGGCGCTGCCACGCCCATGCGTGCACCGTACAAGCTGCCG **************************************************.......................................(((((...(((((..........)))))...........(((....))).........................................((((.......))))..)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
M061 embryo |
SRR060665 9_females_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060668 160x9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................CTCTCTCTTTTCTCGCCGGCTGC.............................................................................................................................................................. | 23 | 3 | 3 | 24.00 | 72 | 72 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TCTCTCTCTTTTCTCGCCGGCT................................................................................................................................................................ | 22 | 3 | 11 | 7.27 | 80 | 80 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAATATAACA................................................................................. | 26 | 0 | 2 | 2.50 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AACAACAACAACAACAAATATAACA................................................................................. | 25 | 0 | 2 | 1.50 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AACAACAACAACAAATATAACA................................................................................. | 22 | 0 | 2 | 1.50 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................CGCTGCCACGCCCATGCGTGCACCGTA.......... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AACAACAACAACAAATATAAC.................................................................................. | 21 | 0 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................CTCTCTCTTTTCTCGCTGGCTGC.............................................................................................................................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGCTATGCGGGCGCTGCCAC............................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CATGCGTGCACCGTACAAGCTGC.. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GCCCATGCGTGCACCGTAC......... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................AAAACAACAACAACAACAAATATAACA................................................................................. | 27 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAATATAAC.................................................................................. | 25 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ACAACAACAACAACAAATATAACA................................................................................. | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAAACAACAACAACAACAAATATAAC.................................................................................. | 26 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ACAACAACAACAAATATAACA................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAATATAACC................................................................................. | 26 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................ACACCAAAAACAACAACAACAAC........................................................................................... | 23 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................TCTCTCTCTTTTCTCGCCGGCTG............................................................................................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CAACAACAACAAATATAAC.................................................................................. | 19 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAAACAACAACAACAACAAATAT..................................................................................... | 23 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AAACAAAAACAAAACACACC............................................................ | 20 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AACAACAACAACAACAAATATAA................................................................................... | 23 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AAACAACAACAACAAATATAAC.................................................................................. | 22 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAATA...................................................................................... | 21 | 0 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAACAACAACAACAAATATA.................................................................................... | 20 | 0 | 14 | 0.14 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ACAACAACAACAAATATAA................................................................................... | 19 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................TCTCTCTCTTTTCTCGCGGGCT................................................................................................................................................................ | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAACAACAACAACAAATAT..................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAAT....................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ACAACAACAACAACAAATAT..................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AAACAACAACAACAACAAA........................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CTTTGTTAAATAAACGAGATGATTAGTGTACATTATATAAGGTAGAGAGAGAGAGAGTGGAAGAGAGAGAGAGAGAGAAAAAAGCGACAGACGTAAGTACGTTGAGAAAGGTACGTAAGGTAGAGAAAACGTGTGCGTGTGGTTTTTGTTGTTGTTGTTGTTTATATTGTGTTTGTTTTTGTTTTGTGTGAACAGAACGTCACTCGATACGCCCGCGACGGTGCGGGTACGCACGTGGCATGTTCGACGGC
**************************************************.......................................(((((...(((((..........)))))...........(((....))).........................................((((.......))))..)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
V047 embryo |
SRR060663 160_0-2h_embryos_total |
M061 embryo |
V053 head |
SRR060678 9x140_testes_total |
M047 female body |
SRR060670 9_testes_total |
SRR060659 Argentina_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060674 9x140_ovaries_total |
SRR1106719 embryo_8-10h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................ATTAGTGTACATTCT....................................................................................................................................................................................................................... | 15 | 1 | 9 | 0.67 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................AAAAAACATGTGCGTGTGGTT........................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CCTCACTCGATGCGCCCG.................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CAAGGTACGTGAGGTAGA............................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACTCGATCGGCCCGCGCCG............................... | 19 | 3 | 19 | 0.21 | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...................................................................................................................................................................................TGTTTTTAGTGATCAGAACGT................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................AGGTGACGCGATACGCCCG.................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TAAGGTAGAGGACACGTG...................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TAGAGAGAAAGAGAGTTGAAGT........................................................................................................................................................................................... | 22 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................CCGGCGACGGTGCGTGTGCGC................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GAAGAGAGAGAGAGAGAGAAA........................................................................................................................................................................... | 21 | 0 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................AGAGAGAGAGAGTGGAAGG........................................................................................................................................................................................... | 19 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................CCGTAAGTACGTTGA.................................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:4903396-4903646 + | dvi_8939 | GAAACAATTTATTTGC---TCT------------------------ACT-A--ATCACATGTAATATAT----TCCATCT--------CTCTCT--------CTCTCACC----------------TTCTCTCTCTCT----------------------------------------------------------------------------------------------CTC---------TCTTTTTTCGCTGTCTGCATT--CATGCAACTCTTT--------------------------------------------------------------------------------------CCATGCA------TTCCAT-------------------------------------------CTCTTTTGCACA---CG---------------C--------------------------------------------------------------------------------ACACCAAAAACAACAACAACAA-------------------------------------------CAAATATAACACAAACAAAA-----A-----CAAAA-----CACA-------------------------------------------------------CT----------------------TGTCTTGCAGTGAGCT-------ATGCGGGCGCTGCCACGCC--CATGCGTGCACCGTACAAGCT------------------------------------------------------------------------------------------------------GCCG |
| droMoj3 | scaffold_6473:113842-114210 - | CAATTTG----TTTGC---TCT------------------------ACT-AAAATCACATGAAATACAT----TCGCTCTCTCTCTCT--------------CTCTCTCTCTATT----CATA------TCATA----------------CACTGTTTCTTCTTGCCTCTCTGTTCCTCTCCTTCGTCATATCGGACATTGTTTGTCCT----------------------TCTC---------TTTC-------TGTCTGC--C--CATATCCATCTCT--------------------------------------------------------------------------------------CACTCTCTTTCCCTTTGGTC-TATCTCCTTCTCTCTCCAACTGTCTC--TCCCT----CTCTGTCTTT--CACA---CGCAC------------------------TCGAAT----------------TGCAACCACATCA---------------------------------ATCAATCACATCAACATCCGAAA----------C-GATCGAC-------------AT-------------------------AAAAACAAAC-----CCGAT-TCCCCACA-------------------------------------------------------CT----------------------TGTCTTGCAGTGAGCT-------ATGCGGGCGGTGCCACGCC--CATGCGTACGCCGTTCAAGCC------------------------------------------------------------------------------------------------------GCAG | |
| droGri2 | scaffold_15081:2759080-2759295 - | TTTG-------TTTGC---CTT-TGTA--------------------CT-A--ATCACAAGCAATATGT----TCTGTCT--------CCCTCT--------CGCTCCCC----------------CACTCTAACCCT-----------------------------------------------------------------------CTCT-------------------CTC---------TCTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCAT----CACTGCATTC--CATG---ACTAC------------------------TTGCAA----------------CACAATG---CACAACGATTAA------------------------AACAACAACAACAATAC-----------------------------------------------------------------AACAAAAAAC-----CCAAC--ATCCAAC-------------------------------------------------------CT----------------------TATCTTGCAGTGAGCT-------ATCCAC------------C--CTTGCGGACACCCTACAAATT------------------------------------------------------------------------------------------------------GCC- | |
| droWil2 | scf2_1100000004590:4306501-4306731 + | GAATTG-----CAT--CGCC-TTTGCTCC-------T--CTCTAATACT-A--ATCACAGGTGATT--TTCCATTGCT--------CT--------------------------------------------CTCTTTCTCTCTCCCCCT-----------------------------------------------------------G--T-------------------TCCACATCC--------------------------------------T--------------------------------------------------------------------------------------TACTTTT------TT-----------------------------------------------CCTCTC--C------------------ATAAAC--ATAAACAA---------------------------A-----ACA-------------------AT------------AAAAACTACATAA--------------------ACACTTAAC-------------ACAATCA-------------------CCAC---CTACACCTGCCAAA-----TCTC-------------------------------------------------------CT----------------------TATATTGCAGTGAGCT-------ATGCAAATG---CCACGCC--TATGCGG---CCCTACATGAT------------------------------------------------------------------------------------------------------GACA | |
| dp5 | XL_group1e:10152943-10153217 + | TTCCCTTTC--T-------CTCT---------------------------------------------------------------CT------------------CTCTCTG----------------TCTCT---------------------------------------------------------------------------TTCTATCTACCGATGCCTGTTGTCTC---------TTTCTATTCAATGCATATCTC--------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCC-TGAACATTCCACATGCCACATGC------------------------ACCACATACAACAA----------------ACAATCGACCAA-----------CAAATGACATTTGCAACCAACAAAAC-------------------TCGAAACCTAC-----ACGAAACATCCTGT-------------------------------------------------------TTTTTTTCCCTCCCGTTCCTTGCGTCTCTTGCAGTGAGCT-------ATGCCGGCGGCGCTGCAGGTCCAGGTGC----------AGGT-------------------------------------------G------GTGTT-------------------------G--------CCCTGCCGGTGGTGCGT | |
| droPer2 | scaffold_32:14098-14326 - | TCCA-------TCT--CTCT-CT---------------------------------------------------------------CT--------------CTCTGTCTCTG----------------TCTCTCTC---------------------------------------------------------------------------T-------------------CTCTCATCCTTCCTTTGAATTGGAATCAC---------TCAGGTCT--------------------------------------------------------TGAAGCAAGAACCCGTTCTCTTAC--------------------------------------------------------------------------------CA---AG--------------------------------------------------C---------------------------AC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGAT-----ACGAGGGAGAAGAAGTAAATC--AA------AACAGCAGAAACACACAAAACCAACTCGACACCAATGAAAAATGTCACAAAGAGGTAAGCGCGGTAATC-------------AACATCATCATCAGTCGCATC--------ATCCGCCG | |
| droAna3 | scaffold_13117:2410540-2410696 - | CACC-------TCT--CTCT-CT---------------------------------------------------------------CT--------------CTCTCTCTCTC-------------------------------------------------------------------------------------------------------------------------------------T---------------------------------------------------------------------------------------------------------------------------CTC------TCTGGTGCTATCT--------------------CTTGC--TCCCTC----A----------C-------------------------------TTT-C-----------------------------C---------------------------------CGCCGAAACAAAGACAA-----------------------------------------------------------------ATCGACCAAC-----CCGAC-----TCCC-----------------------------------------------------C-TT----------------------TTACTTGCAGTGAGCT-------ATGCGGGCGCCGCCTCGCC--CATCCGC---CCCTACAAGGT------------------------------------------------------------------------------------------------------GACC | |
| droBip1 | scf7180000396359:119476-119708 - | AATATTTTC--T-------CCCT---------------------------------------------------------------CTTTCTCT--------CTCTC-----G----------------TCCCTCTC---------------------------------------------------------------------------T-------------------TTC---------TTTCTGTTT---------------------------------TTTT------------------------------------------------------------------------------------------------------------------------------------------TCTTTC--C------------------ATGCCATGCC-----ACCGCAAC----------------TGCA----------ACTACTGCAACAACTACTGCAA-----------------ACA--ACAATGCAAACTA-----------CAGAGCGCAATTGTAAGCGAAATGAG-------------------CACAGAATTGT-----GAGAATTAGCTAAA-------------------------------------------------------TT----------------------TAATTTGCAATAGCCA-------AAGCCCGCCGCCCACAACGACCACGCCCATGCAGTAACAGCT--------------------------------------------------------------------------------------------------------GG | |
| droKik1 | scf7180000302621:180849-181042 - | TCTA-------TCT--CTCT-GT---------------------------------------------------------------CT--------------TTCTTCCTCTATA----TATAACTCTCTTTCTCTCT----------------------------------------------------------------------------------------------CTC---------TCTCTATCTC-TGACTATGTG--TGTGTATCTCTCT--------------------------------------------------------------------------------------TTCTCTC------TCCCGT-------------------------------------------GTCTCCCA-----CAA---------------TT--ATTGTTTA---------------------------T-----ATA-------------------AA------------AAAAAACA--------------------------------------------------------------------------------------------------------------------------------------------------------------CA----------------------TATTTTGCAATTGCCTCAGATCAACGGCAGCGGAGTTGCAGC--TG------CACAGCAGCAACA------------------------------------------------------------------------------------------------------GTCG | |
| droFic1 | scf7180000453882:157390-157632 - | CTCT-------CCT--CACC-TCTCC-----------------------------------------------------------------------------TCTTCTCTTC----------------TCTTTC-----------------------------------------------------------------------------TT-------------------TTTTCTTCCCCTTTTGGTTT---------------------------------TGTTTCTTTCG-------------C----TTT----------------TCTCGTTGGCAGCTGCCGTTCGGCGAAGCCCAACAAGCG------TTGCAG-------------------------------------------C------------TA-----------------C------------------------------------CAGCGGCAAC---------------------------------AGTAGCAGCAGCAGTAGCAGCAG-------------------------------------------CAGCAGCAACAACATCAGCA-----G-----CAGCA-----GCAG-------------------------------------------------------CA----------------------GCAGCGGCGGCAAACG-------CAACAGCAGCAGCAGCGGC--ACCGCAACTACCGCCCCCGC------------------------------------------------------------------------------------------------------TGCCC | |
| droEle1 | scf7180000491043:57660-57868 + | TCTC-------C----------------C------------------------------------------------------------------------CATCTCGCCCTG----------------TTTCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTCGATGTCTTCCCG--CATATACACCTCTCGCTCTCATTTGT--------------------------------------------------------------------------------------TTTAAT-------------------------------------------GTTCCATT-----T--------------------------------GCAA----------------TCCAA------------------------------------------------ACATTACAATCCAAAC-----------AACCGACC-------------GCAAAAATCGG----CA----------ATCAACCAAC-----CCGAATCGAATCCC-------------------------------------------------------TT----------------------TACCTTGCAGTGAGCT-------ATGCGGGCGGAGTGCCACC--CATTCG---------------------------------------------------------------------------------------------------------------------GCCG | |
| droRho1 | scf7180000779333:867218-867437 - | TCTT-------TCT--TTCT-CC-----C------------------------------------------------------------------------CATCTCCCTCTG----------------TTCCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTTGATGTCTTTCCT--CATACGCACCTCT--------------------------------------------------------------------------------------CACTCAC------CTCGGT-------------------------------------------GTCATGTA-----CT-------------------------------------------------------------TTA---------------------------------ATTTACTACATCATAATCCGAACAA---C--AACAACCGACC-------------GCATAAA-------------------CCGGAACCAAC-----CCGAA-----TCCC-------------------------------------------------------TT----------------------TACCTTGCAGTGAGTT-------ATGCGGGCGGAGTACCACC--CATTCGC---CCTTTTAAGGT------------------------------------------------------------------------------------------------------GGC- | |
| droBia1 | scf7180000302421:1620354-1620586 - | TCTT-------TCT--CTCT-TT-CTCCC-----------------------------------------------------------------A------CATCTGCCTCTG----------------TCCCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTCGATGTCCTCCCG--CGCACACCCCTCT--------------------------------------------------------------------------------------CACTCAC------TCCGGT-------------------------------------------GTCCAATA-----TA-----------------C------------------------------------AAACATATCA---------------------------------ACTAACCACATCATTATCCGAACA-----------AACCGAC-------------GCA-AAA-------------------CCACAACCAAC-----CCGAA-----TCCC-------------------------------------------------------TT----------------------TGCCTTCCAGTGAGTT-------ACGGGGGCGGAGTGGCGCC--CATTCGC---GCCATCAAGAT--------------------------------------------------------------------------------GGCCGGAG----------------CT | |
| droTak1 | scf7180000414531:38474-38778 - | ATTTG------TTTAT---ATAT-ATA------TTACATATATATT----T------------ATA-----------------------------------TATATATTTTCCTTT-TTTTTGTCTCTTTTTCTCTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTTT----CCGTGTTTTT--CAAA---ACTTCCTGTCCGATGCACTGCT-----ACAAA-A---------------------A-----ATA-------------------------------------AA------------TACAA--ATACATAAATGCACCAC-------------ACA----CCGA---------------CAACAAAAAAA-----ACGAAAAATGTATACCCACAACACTGTGAAAAAAACAGTCGCAGTTCCGATTCGACAACGAGTGCTATCTC----------------------AATCTGCCGGACAATC-------ACA---------------------------------------------------------------------------AGCGGGTGAG--------TCACTTTCAACTTCTGCCAGCGACCTTGGGGGCGTC--------AT-CACTG | |
| droEug1 | scf7180000409110:1554615-1554848 - | TTTT-------TCT--TTCT-TTTCCT-C------------------------------------------------------------------------CATTTGTCTCTGTT---------------TTCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCAATCGATGTCTTCCTGTTCATATACACCTCT--------------------------------------------------------------------------------------CACTCAC------TTTGGT-------------------------------------------GTCATATA-----TA-----------------T----ATA------------------------------TATATATAT---------------------------------ATTAACTACATCATTATCCGAACAA---------AACCTGCC-------------ACAAAAA-------------------CCACAACCAAC-----CCGAA-----TCCC-----------------------------------------------------TCTT----------------------AACCTTCCAGTGAACT-------ATGCGGGTGGAGTTGCCCC--CGTTCGG---ACTTTTAAGGT--------------------------------------------------------------------------------------------------------GG | |
| dm3 | chrX:15844188-15844394 - | GATAAAGGTTGGTTGC---CCC-TCTAACTAGCTAAC------------T-----------------------------------------------------------TATG----------------TAACT---------------------------------------------------------------------------TACTA-------------------CCATCTTGATCTTGTGATCT---------------------------------TATTTCTTAAGGGTCTTGACTTCACATCGTTTTCCTATTTACTATTAC----------------------GT-----------------------------AATCA--------------------CTTGC--ACATCT----C----------T-------------------------------TTT-T-----------------------------A---------------------------------AACAACAACAACAACAACGACAA-------------------------------------------CAACTAAAACAACAACAACAACAAC----------A-----ACAA-------------------------------------------------------TT----------------------TGCATTGTAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | x:15015898-15016112 - | GATAAAGGTTGGTTGC---CCC-TCTAACTAACTCAC------------T-----------------------------------------------------------TATG----------------TAACT---------------------------------------------------------------------------TACTA-------------------CCATCTTGATCTTGTGGTCT---------------------------------TATTTCTTAAGGGTCTTGACTTCACATCGTTTTCCTATTTACTATTAC----------------------GT-----------------------------AATCA--------------------CTTGC--ACATCT----C----------T-------------------------------TTT-T-----------------------------A---------------------------------AACAAC------AACAACTACAA-------------------------------------------CAACTAAAACAACAACAACAACAAC------------------AA-------------------------------------------------------CT----------------------TGCATTGTAATA------------GA------------------------T---GCGTTGACATT--------------------------------------------------------------------------------------------------------GC | |
| droSec2 | scaffold_21:840614-840838 - | CCGT----------------------------------------------------------------------------------------------------CTTCCTCTGTTTAAATCCGACTGCCTTCCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTCGATGTCTCCCCG--TATATACACCTCT--------------------------------------------------------------------------------------CACTCAC------TCTGGT-------------------------------------------GACGTGTA-----TA-----------------C---------------------------------------CTAATTA---------------------------------CCGAACCACATCATTATCCAAAA----------CAACTTGCT-------------GCAAAAA-------------------CCACAACCAAC-----CCGAA-----TCCC-------------------------------------------------------TT----------------------TACCTTCCAGTGAGTT-------ATGGGGGCGGAGCTTCGCA--AATTCGC---ACCTTAAAGGT-----------------------------------------------------------------------------------------------------AGCCG | |
| droYak3 | X:7365391-7365657 - | GAAACTCTT--TCT--ATTT-TC-TTC-C---------------------C-------------------------------------------ATCTTTCTATCTTCCTCTGTTC---TCCGACCGCCTTCCTCTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTCGATGTCTCCCCG--CATATACACCTCT--------------------------------------------------------------------------------------CACTCAC------TCTGGTG-TAT--------------------------------------GTCGTGTA-----TG-----------------C---------------------------------------CTAATTA-------------------------ACCAACCAACTAACCACAACATTATCCAAACAA---CCAAACAACTTGCT-------------GCGAAAA-------------------CCACAACCAAC-----CCGAA-----TCCC-------------------------------------------------------TT----------------------TACCTTCCAGTGAGTT-------ATGCGGGCGGAGCTGCTCC--AATTCGC---GCCTTTAAGGT--------------------------------------------------------------------------------------------------------GG | |
| droEre2 | scaffold_4690:13780891-13781128 + | CTCTT------TCT--C--T-TTTCTTCC------------------------------------------------------------------------AATCTTCCTCTGTTT---TCCGGCTGCCTTCCCTTGT----------------------------------------------------------------------------------------------CTC---------TTTCCATTCGATGTCTCCCCG--TATACACACCTCT--------------------------------------------------------------------------------------CACTCAC------TCTGGT-------------------------------------------GTCGTGTA-----AA-----------------C---------------------------------------CTAATTA---------------------------------ACCAACCACATCATTATCCAAAC-----------AACTTGCT-------------GCAAAAA-------------------CCACAACCAAC-----CCGAA-----TCCC-------------------------------------------------------TT----------------------TGCCTTCCAGTGAGTT-------ATGCGGGCGGCGCTGCACC--AATTCGC---GCCTTTAAGAT-----------------------------------------------------------------------------------------------------TGCCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 05:06 AM