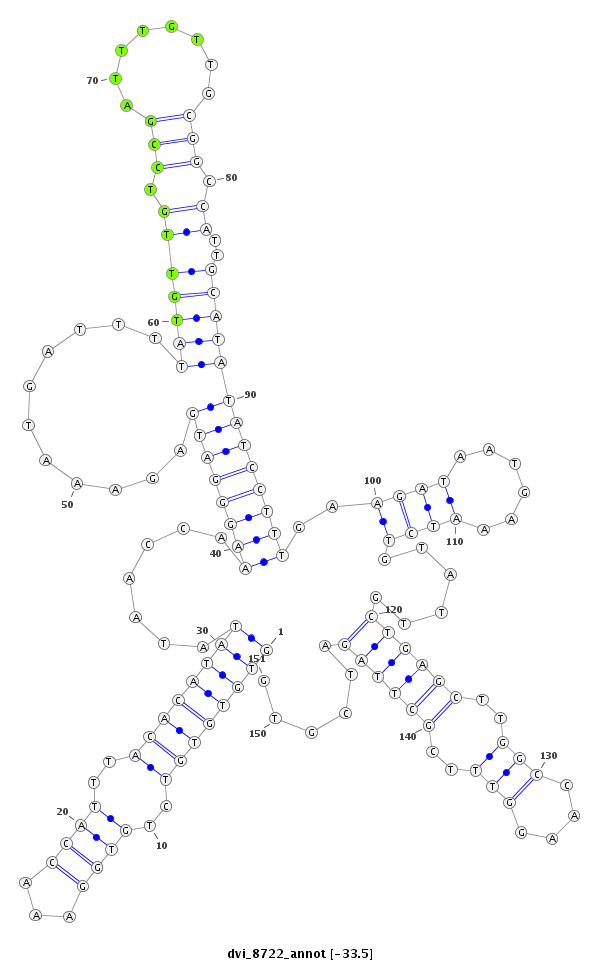
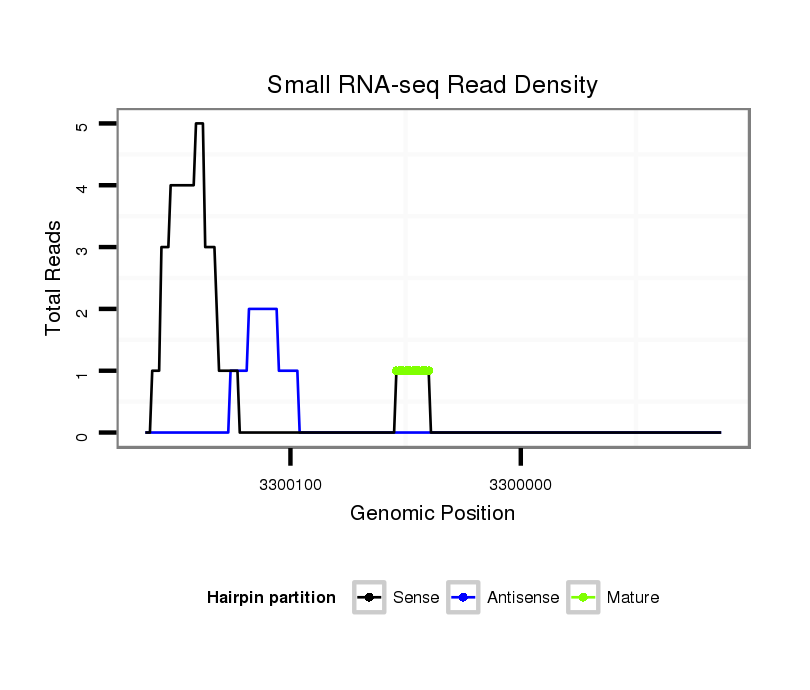
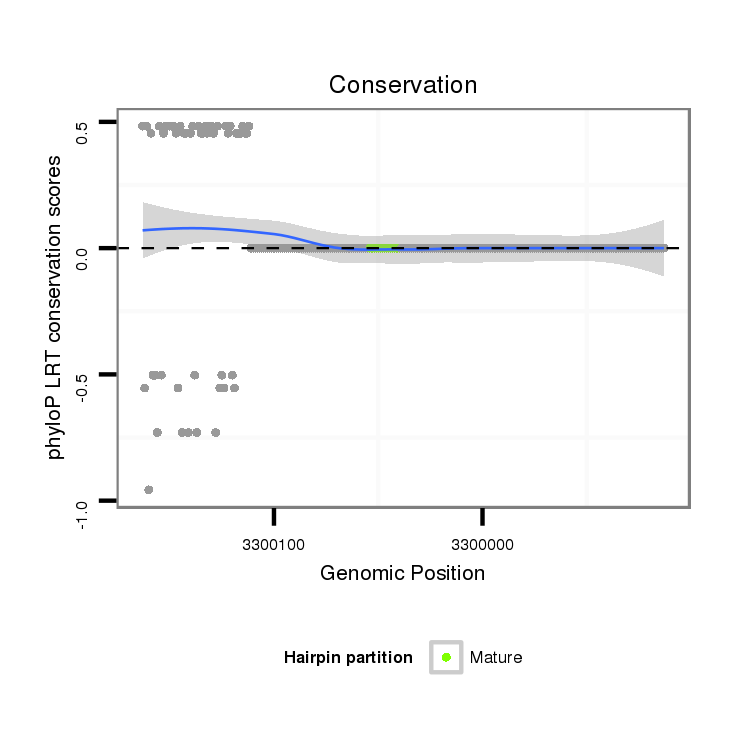
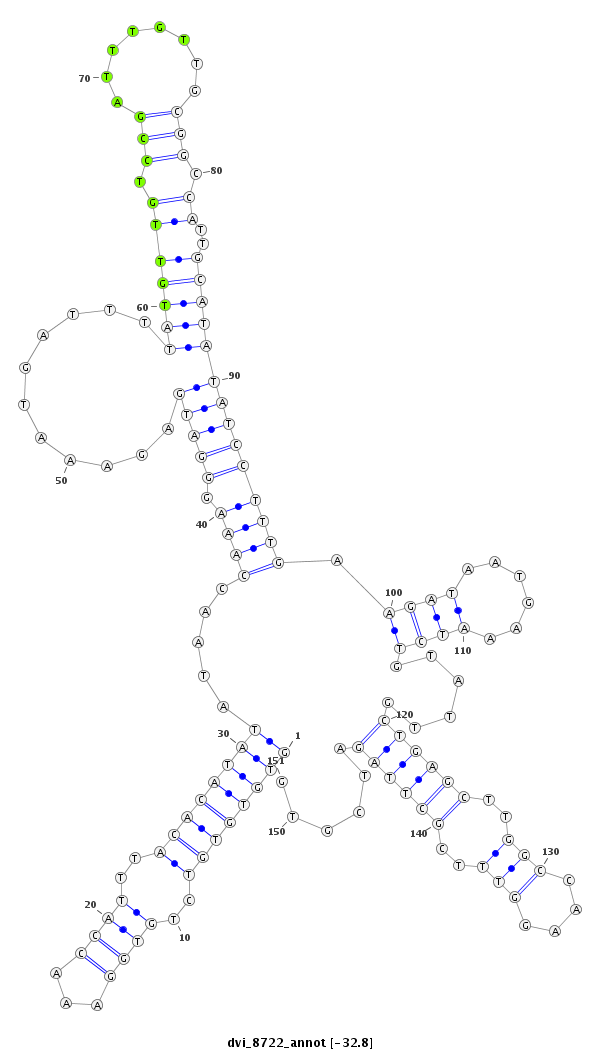
ID:dvi_8722 |
Coordinate:scaffold_12928:3299963-3300113 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.8 | -32.8 | -32.8 |
 |
 |
 |
exon [dvir_GLEANR_17082:3]; CDS [Dvir\GJ16574-cds]; intron [Dvir\GJ16574-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TATCGGAGAGCTTTAAGATTGGAGATCTGCTCAAGCTTTTTGTATCCGAGGTGTGTGTCTGTGGAAACCATTTACACATATATAACCAAAGGGATGAGAAATGATTTTATGTTGTCCGATTTGTTGCGGCCATTGCATATATCCTTTGAAGATAATGAAATCTGTATTGCTGAGCTTGGCCAAGGTTTCGCTTAGATCGTGATTCTTCTCAGGGCAGCCAATGCAAAGTGTCATGACGATAGAGAGATAGA **************************************************((((((((..((((...))))..)))))))).......((((((((...........(((((((.(((........))).))..)))))))))))))..((((......))))......((((((..(((....)))..))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060663 160_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060682 9x140_0-2h_embryos_total |
GSM1528803 follicle cells |
M027 male body |
M028 head |
SRR060681 Argx9_testes_total |
M047 female body |
SRR060671 9x160_males_carcasses_total |
SRR060689 160x9_testes_total |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................GAAAACTGGATTGCTGTGCTT.......................................................................... | 21 | 3 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................CGACTTTTTGTTGTCCGATTTGT............................................................................................................................... | 23 | 3 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TTCTCCGATTTGTAGCGG.......................................................................................................................... | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......GAGCTTTAAGATTGGAGATCTGCT............................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TTTAAGATTGGAGATCTGCTC........................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...CGGAGAGCTTTAAGATTGGAGAT................................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AGATCTGCTCAAGCTTTTT.................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......GAGCTTTAAGATTGGAGAT................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TGTTGTCCGATTTGT............................................................................................................................... | 15 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGATCATTAGAAGATAATGAAA........................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................CTTTTTGTCTCCGAGGTC...................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CTTTGTTGTCCGATTTGTT.............................................................................................................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CTCTGTTGTCCGATTTGT............................................................................................................................... | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ....................................................................................................................................................................................................................................TGTGGTGACGATAGTGAGA.... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................ATATCCCCTTTGAAGATGAT............................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATAGCCTCTCGAAATTCTAACCTCTAGACGAGTTCGAAAAACATAGGCTCCACACACAGACACCTTTGGTAAATGTGTATATATTGGTTTCCCTACTCTTTACTAAAATACAACAGGCTAAACAACGCCGGTAACGTATATAGGAAACTTCTATTACTTTAGACATAACGACTCGAACCGGTTCCAAAGCGAATCTAGCACTAAGAAGAGTCCCGTCGGTTACGTTTCACAGTACTGCTATCTCTCTATCT
**************************************************((((((((..((((...))))..)))))))).......((((((((...........(((((((.(((........))).))..)))))))))))))..((((......))))......((((((..(((....)))..))))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060659 Argentina_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060680 9xArg_testes_total |
SRR060657 140_testes_total |
M047 female body |
SRR060671 9x160_males_carcasses_total |
V047 embryo |
SRR060684 140x9_0-2h_embryos_total |
SRR060683 160_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GCTAAACATGGCCGGTAA..................................................................................................................... | 18 | 2 | 3 | 1.33 | 4 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................GGCTCCACACACAGACACCTTT........................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GCATGTGTATCTATTGGTTTCC............................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................AAAACATAGGCTCCACACACA................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GCTAAACATGGCCGGTAAC.................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................ATGTGGATCTATTGGTTTCC............................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CGAATCTAGAACTAGGAAG........................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................TCTTTACTCTCTACTAAAATAC............................................................................................................................................ | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................AAAATGCAACAGGCTAAGCT............................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................CGGTTAAGTTTCAGAGTA................. | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................TATGTCTATTTATTGGTTTCC............................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................CTTTGTATCTATTGGTTTCC............................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:3299913-3300163 - | dvi_8722 | TATCGGAGAGCTTTAAGATTGGAGATCTGCTCAAGCTTTTTGTATCCGAGGTGTGTGTCTGTGGAAACCATTTACACATATATAACCAAAGGGATGAGAAATGATTTTATGTTGTCCGATTTGTTGCGGCCATTGCATATATCCTTTGAAGATAATGAAATCTGTATTGCTGAGCTTGGCCAAGGTTTCGCTTAGATCGTGATTCTTCTCAGGGCAGCCAATGCAAAGTGTCATGACGATAGAGAGATAGA |
| droMoj3 | scaffold_6308:170009-170060 - | TTTGGAGTAACTTTAAGTTGGGCGACATGCTCAAGATACATGTGACCGAGGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 10:08 PM