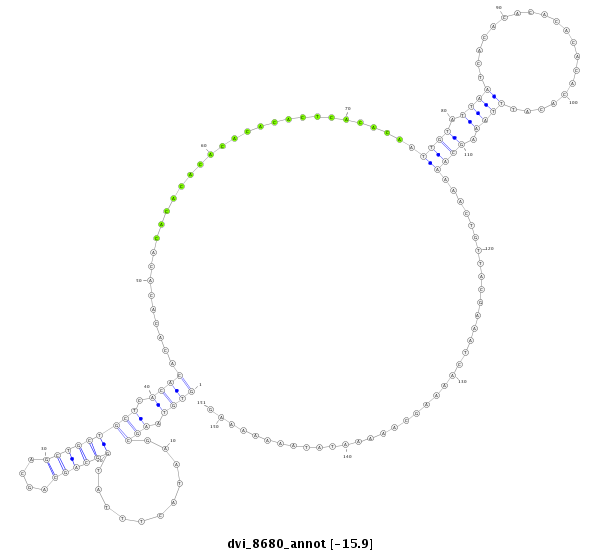
| droVir3 |
scaffold_12928:2778907-2779157 + |
dvi_8680 |
TGATGTGA---------C-----CTGC--CG------CCGCAGCAGCAG------CA-GCAGCAGCAGCAGTTGCAG---------TTGT-----------GTAAGCGAA-----TACTTTATG-G-----------CAGCAGCAGCTGCT------------------GCT-----------------------------------------------------------------------------------------CACACACA-CACACACACACA--------------CACACAC------------------ACTCACACAAT------------------------------------TGTATTAA------TCACACACACACACACACA------T-------TT--AA--------------------------------------------------------------------AGCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACTGTTACGAAATCAAAAG--------C--AA----------------------------AAA-----------------------------------------------------TATAAAAAAAG---------------A---------GCAACTGCTGTGACG-------------------------------------------------------AGGACGAGGACGAGGACGAGAACGT------GGACGT----------GGA |
| droMoj3 |
scaffold_6473:14568805-14569009 + |
|
TGATTTGA---------C-----CTGC--AG------GAGC------AGCAGTTGA---------------------------------------------ATAAACGAC-----TGCTTTATG-GCAAC------G--GCAGCAGCT---------------------GCT------------------------------------------------------------------------------A--CA------CA--CAC-ACACACACACAGA--------------TA-------------------------------------------------------------------------------------------------------TGTATT---------T--TT--------------------------------------------------------------------AGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAACTGTTACGAAATCAACAAAAA---A--ATAATAT------TTATATATAT-----------------------------------------------------------------------------------------------------------------------------------------ATATCTCTGTGTGTGTGTGT------------GTGTGTCTGTGTCCAGGAGGTGGTCTGAAAAGGGGAAAA----------AAG |
| droGri2 |
scaffold_15203:1678244-1678475 - |
|
TGATCTGA---------C-----ATGC--CA------CAGCAGCAGCAG-------------CAGCAGCAGTTGTAG---------TTGA-----------ACAAGTGAC-----AGCTTTATG-G-----------AAGTAGCAGCTGCC------------------GCA-----------------------------------------------------------------------------CTAGCACATA------AAC-----ACACAAACA--------------CACTCAT------------------ACTATT-----------------------------------------------AA------TC----------------G------TCA-------------------------------------CA---------------------------------------TGGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACTGTTACGAAATAAAAAAAAA---A--AACAAAA------TAATAAAAAT----------------------------------AAG----------------------------GAGAGAAAAAG---------------A---------GCAACTGCTGTGACGAGGAC------------------------------------------------------------C------------GA------GGACAATGACTGTGGTGGG |
| droWil2 |
scf2_1100000004768:764510-764766 - |
|
TGCCGACA---------A-----CAGC--AG------CAGC------AGCAGCAGCA-ACAGCAACAGCA---ACAG------CAGCA-------------------------ATAACATCATA-G---CCTGCGC-TCCTTTTGGTGGCCAG----------------------------------------------------------------------------------------------------CA------CACACACA-CACACACACACA---------------------------------------------------AA----CTGAGACAC-------ACATACAGTGAGGAGAGATAATAACAATC----------------G------ACA-------------------------------------CA---------------------------------------C-------------------------------AGTCGGGCCAAGAACCAAGCAGTAAGACCGACCACAAAGCAGACCAACCAACAGCAACAGCAACAGCAAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---------GAAACAGCCGAGA-------------------------------------------------------------------------CCAGCGCAA------AGACCA------G---GGC |
| dp5 |
XL_group1a:2070257-2070424 - |
|
GCAGC----------------------AG--CAGCAGCAG------CAG----------CAGCAGCAGCAGTAGCAG------TAGCACTC-------------C-----CCAGTAACCGTAATCGTAGC------C--TTAACAGTAACT------------------G------------------------------------------------------------------TTAACACCCATTTCCGCCAC------TCAC-----ACACA----------------------------------------------CACACGCGAG------------------------------------TGCG-----------CACACATACACACAAACA------A-------AA--CA--------------------------------------------------------------------GGAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACTGTTCCCCAAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CA |
| droPer2 |
scaffold_22:852790-853023 + |
|
AGCAGC---------------------AT--CAG---GG------GCAG------CA-GCAACAACAGGA---AGA--------------------------CAT-------------------------------------------------------------------AACA---CACATCCTTTGG--------------------------------------------------------------CA------CACACACA-CACACACACACA---------------------------------------------------CA----CAGACACACCCCG-AAGCACACTCCG-----------------------------------------------------------------------------------------AC--------------------------------------------------------------------------------------------------------------------------------------AAATCTAATAATAAATCCAAG---------------------------------------------------------------------ACACTTAACGCCTT----GGCCGCAG------------------------------------------------------------------CAACCATGACCCACAAACTAGCTGAGGGATTGCCCGAATG--------------------------------------------------TGCTGTGTGC----------------------------------------T------------GGAAGCGGAAGGAGTTGGAGTGTTCGA------GGACGT----------GGA |
| droAna3 |
scaffold_12903:139958-140172 + |
|
TGCAGT---------------------TG--CAA---GG------ATAA------CG-GCAGGACATGCA------G------CCAG---CAACAAGTGAGACAACCGCA-----TAATCCACA-G-----------TAACCGCAAAGGCC------------------ACG-----------------------------------------------------------------------------CCCACACACA-----------CACACACACACA--------------CCCGCAC------------------ACCCACACACA------------------------------------AATAAAAA------CCACTC-------------------TCACACACACGAGAAGCCGCATTAATAAATTTTCATGGTAAAAAAAAAAAGTGGAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAT-----------------------------------------------------------------ATTG------------------------------------------------------AAAAA------GCGATC----------TCT |
| droBip1 |
scf7180000396425:706815-707038 + |
|
GCAGC----------------------GG--CGGCG---------GCGG------TG-GCGGTGGCGGCG---GCGG------TGAC---CCACGAAAAAGGCAAGCACT-----GGAAGAGGA-A-----------GAGCCTGAG-------------------------------------ACCGTTCA----------------------------------------------------------CTCATA----------CAC-ACACATATACACA--------------TA-------------------------------------------------------------------------------------------------------TACAGT---------G--TT--------------------------------------------------------------------ACTG--------------------------------------------------------------------------TTACTGGTAATGGCTACTGCCAC-----------------------------------------TGGCA-------------------------------------------------------------CACGCAAAA------------------------AAT--AA----------------------------AAATAATAA----------------------------------------ATAGGAATATAGAAGTAC---------------G---------AGTACGCCT-------------------------------------------------------------------------GCGACTGTGACTG------CGACTG------C---GAC |
| droKik1 |
scf7180000302570:78819-79054 + |
|
TGCAACAA---------A-----CTGC--AG------CACCAGCAGCAG------CA-GCAGCAACTGAA---GTGG------CGGT---CCA------ATGCATGCGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------TGAAATTAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACTGAAGTTTTGCACGCTAC-----------------------ATTTAAAGCAG-------CAGCAAACGGCAAATGCAACAGCAAC-----------------------------------------AAGAAGA--------------------------------------------AGGAAAAGCCTG----------ACCTGCAACAATGTGAAAATGCA---A--ACTGCGA------TCATAAAAATTGTGG------------------------------------------------------------CCCAA-----CTGCAACCGCTGCCAA----------------------------------------------------------------------------------------------AAC-GGGACGT------GAACGT----------GGA |
| droFic1 |
scf7180000454073:1081245-1081439 - |
|
TTACGTTGCCGGACCGCCGATGACGAC--AACAGCAGCAG------CAGCAGCAGCA-GCAGCA-CAGCA-------------------------------------------------------GCGGC------A--GCGACAGCAGCA------------------G-----------------------------------------------------------------------------------CA------CACACACA-CTCGCACACACACATCGACAACCACACTCACTCACACACACACTTTCGCACAC---------TCACACAC---CACACAGAATAGCAA-----------------------------------------------------------------------------------------AAAAAAAAGAACGCAAAAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTG------------------------------------------------------AAGA------------------------AG |
| droEle1 |
scf7180000491347:8096-8303 + |
|
TGTTTTAA-----TT--A-----CTGA--AG------CAACAGCAGCAG------CA-GCAGCAACAGAA---ACAG------CCACA----------------------TCTCTGGCATTGCA-C-----------AAGCCGCAGCC---------------------GAA-ACAAAACGGAGCCAGCCACACAGACC------------GAGCAAAAGAAATTGAAAAAATGTAATAAAATAAATTTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTAAC-------------------------------------AAAACAAAAATTAAAAAGCAGAAAT--------------------------------------------------------------------CAA---------------------------------------------------------------------------------------AACAAAACAAAACAACGG------CGCAGC----------GAC |
| droRho1 |
scf7180000779479:322601-322783 - |
|
AGAACAGC---------CAGAAGTTGC--AG------TTGCAGCAGCAG------CA-GCAGCAGCAGCAGCTCCAG---------TTGCC-------------A-----CTAGCAACATCAAGAGCGGC------A--GCAACAGCAGCA------------------G-----------------------------------------------------------------------------------CA------T--------------CAATCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACCGGCAACAATAGCAGTCAGCAGGACG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGG---------------A---------GGAACTG--------------------------------------------GACGA------------GGATGAGGAGGACGAGGAGGAGGACGA------GGACGA----------GGA |
| droBia1 |
scf7180000302126:3412145-3412376 - |
|
AGCAGCAA---------C-----TTGC--GG-CC---GAGCAGCAGCAG------CGAGCGGCGGCAGCGGCGGCCG---------TAGT-----------GGCA-------------------------------------GCGGCGGTCAGTGCGCTGCACAACGGC---AACA---AGCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGCACTACAACAACAACAA----CAAC------------------AATAACA-------------------------ATGGCAAC------AGC----------------------------------AAGTCGAGGGATCGCGACGATCACCTGCAGGCACAG-----CAGCAGTCGC------AGCAGTCGCAGCA-----------------------------------------------------------------GCCGCAGGACGACGAGGACGAGGACGA------GGACTA----------CGA |
| droTak1 |
scf7180000415876:134091-134317 + |
|
CCGACC----------------------G--AGGCAGCAGC------AGCAGCAG----------CAACAACAACAG---CAGCAACCAG-----------CGAAG----TGGCTGGCAAAGCA-A-----------ATGGTGAAATC-------------------------------------------------------------------------------------------------------------------------------------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAAAAACATTGGCTGCCAGG--GAAGAACAAGAAAATGGCGCCAATTTGGAGCAAGAAACCAAAAA-----------------------------------------------------------TAC------CAATAAGCCA-----------------------------------------------------------------------------------------------------------------------------------------ATAATACCGAGTGTAAATGAAGAGGAGGACGAGGAGGAGGAGGAGGAGGACGACGACGA------GGAGGA----------GGA |
| droEug1 |
scf7180000409032:624221-624356 - |
|
GCAGC----------------------AG--CAGCA---------GCAG-------------CAGCAGCAGCCGCCA---------TAGT-----------AC-A-----GAAGCAACATCACA-G------------C---------------------------------AACA---TGCAACCTGCAA-----------------------------------------------------------CACCAGCAGCATACTAAC-----ACACACTCA---------------------------------------------------CA----CCGACACAC-----AAGCATACACTGAACTGAACTAA------C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TA-----------TA |
| dm3 |
chrX:18080702-18080897 + |
|
CAAACACA---------A------TGG--GG------CAGC------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACACACG-TGCACACGTACA--------------CACACAC------------AC----AC--ACACACA-CA--------------------------------------------------CACACA----------CACACA---------T--GC--------------------------------------------------------------------ACATATACTGGAAGAGTCAAGTCTGACA---------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTAACGACTTACTTAGCCAAAG------------------------AAC--CA----------------------------AAA-----------------------------------------------------AAAGAAGAAGG---------------G---------GCAGCAACGGTTCCAAGAACCCAAAGAAAAATGCTG------------------------------------------------------AAAAG------GTACGA----------GGA |
| droSim2 |
2l:19896177-19896323 - |
|
CTGC-------------------------AG------CGGCGGCAGCAG------CA-GCAGCGGCGGCG---GCGG------CTGTGGAC-------------G-----TCGATGGTTTTGTG-G-----------ACGACGTTGTGGCT------------------TCT-----------------------------------------------------------------------------CCGCTACCCG------G-----ACGCACTCACA--------------GGCTCAC------------------GCTCACGCACA-------------------------------------------------------------CGCAGACGCACACTTGGCACACTT--AA--------------------------------------------------------------------AG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CA |
| droSec2 |
scaffold_8:204327-204515 - |
|
TGCATT----------------------A--TTGCCGCTGC------TGCTGCTG----------CTTCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CA--CAC---------------ATTAACATTAATCGTCATGCAAACGCACACATACACACAC---------ACACATAG---CACACACGGCACTCA-----------------------------------------------------------------------------------------CACAGCGAGAAAGTTACGAAGCCGAGGGACCAGCAAACGAGC-------------------------------AAA-----------------------ACAAACTACGAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGACGAACGCGACAACGACAACAC-------AGCGA----------CAA |
| droYak3 |
X:19081126-19081375 - |
|
CCGC-------------------------CG------CCGCAGCAGCAG------CA-GCAGCAGCGGCA---GCAG------CGCTCAGC-------------A-----TCAGCAACAACA-----------------------------------------------GC-AACAATTCGGCG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGATCAGCGGGCCAGCAGCCGCAC-------------------------------AGC-----------------------ACTAGCAGCAGCA-------GCA-------ACAAC---------AACAATAACAATGGGGCTAAC---------------------------------------------------------------------ACTGTTAAC------------------AGCAACA-------------------------ACAACAAC-------------------AGCAACAA----------------------------------------CTATGGGCACAG-----CAGCAGCAGCTCGCAGA---------GCAG-------------------------------------------CATGGACGA------------AGAGGAGGACGACGAGGAGGAGGACGA------AGAGGA----------GGA |
| droEre2 |
scaffold_4820:9126974-9127244 + |
|
GCGGC----------------------AG--CAGCAGTAG------CAGCAGCAG----------CAGCA---GCAAGTG---TGA------------------GCCGAA-----TATTCAAAA-A-----------AGGCAGCAAC--------------------------------CGGCGCCAGCCGTCCGTGCCCAAATGGGGATCAAGTGGAGG----------------TGGACGCCAGAACCCCACAC------A--CT---CACACA----------------------------------------------CACCCACAAG------------------------------------GGAGG----------------------------------------------------------------------------------------------------GGGCCAACCGATGCCC-------------------------------TGT-----------------------CCT-----------------------------------------------------------------------------------------------------------------------------------------------------GGCCAAAGCAGCCATCAAAGCACAAG-TCAATGATGCCATTA------TAATAGAGAA----------------------------------------------------------------AC-----------------------------GACGGAGGA-----------------------------------------------------------------GCAGCGCGGAAACGGCGACGAGGACGA------GGACTT----------GGA |