
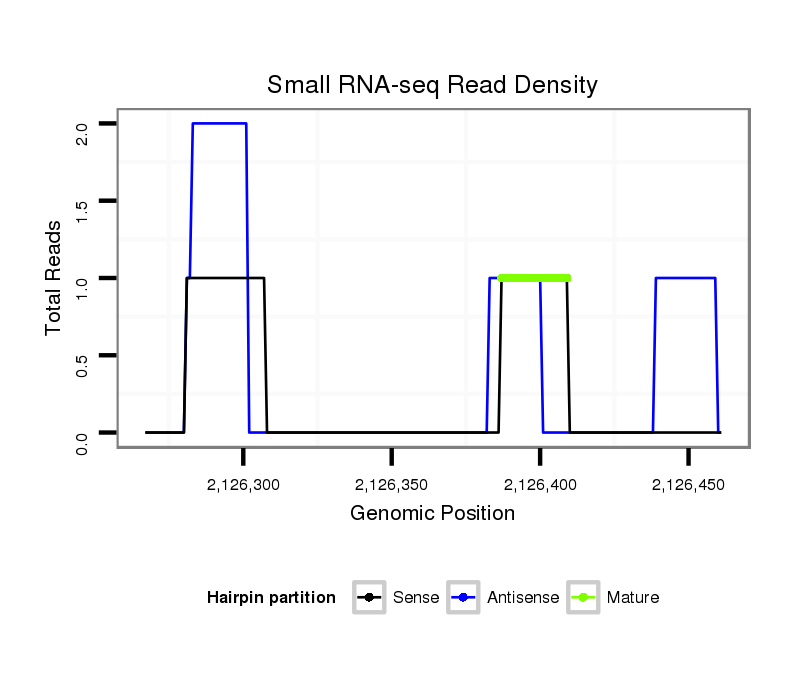
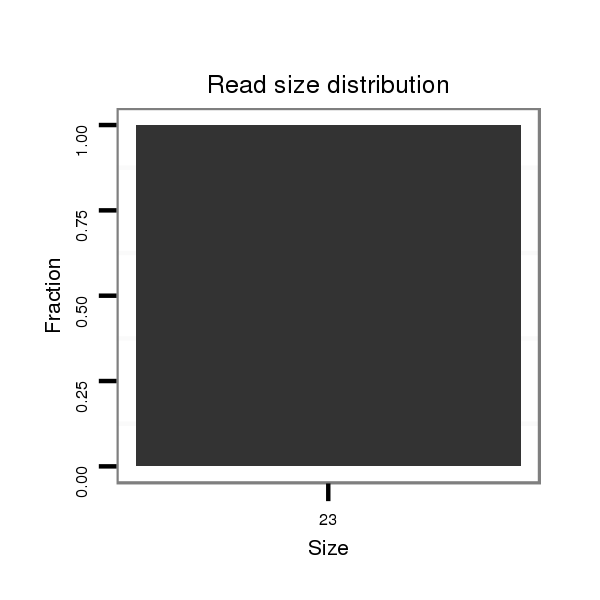

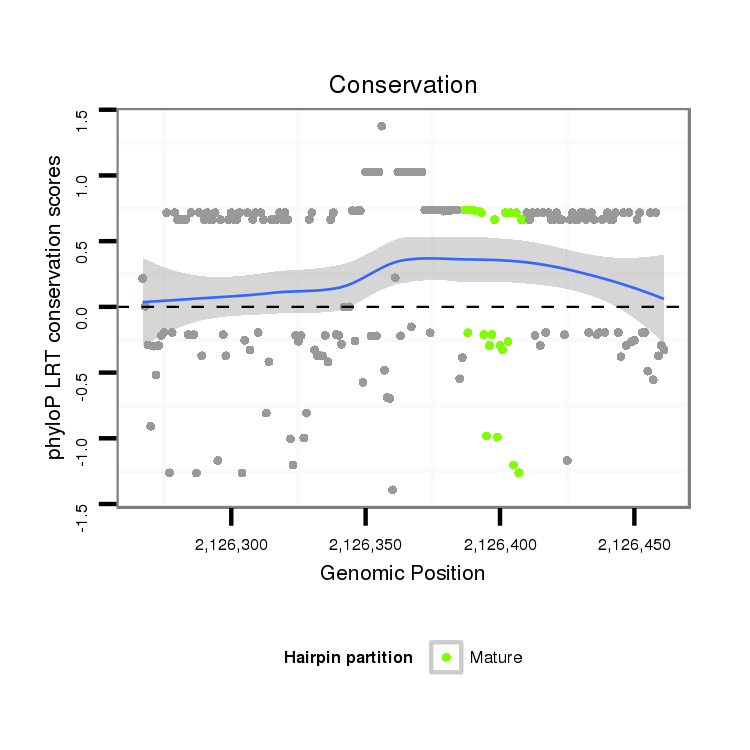
ID:dvi_8588 |
Coordinate:scaffold_12928:2126317-2126411 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.6 | -16.2 | -16.1 | -15.9 |
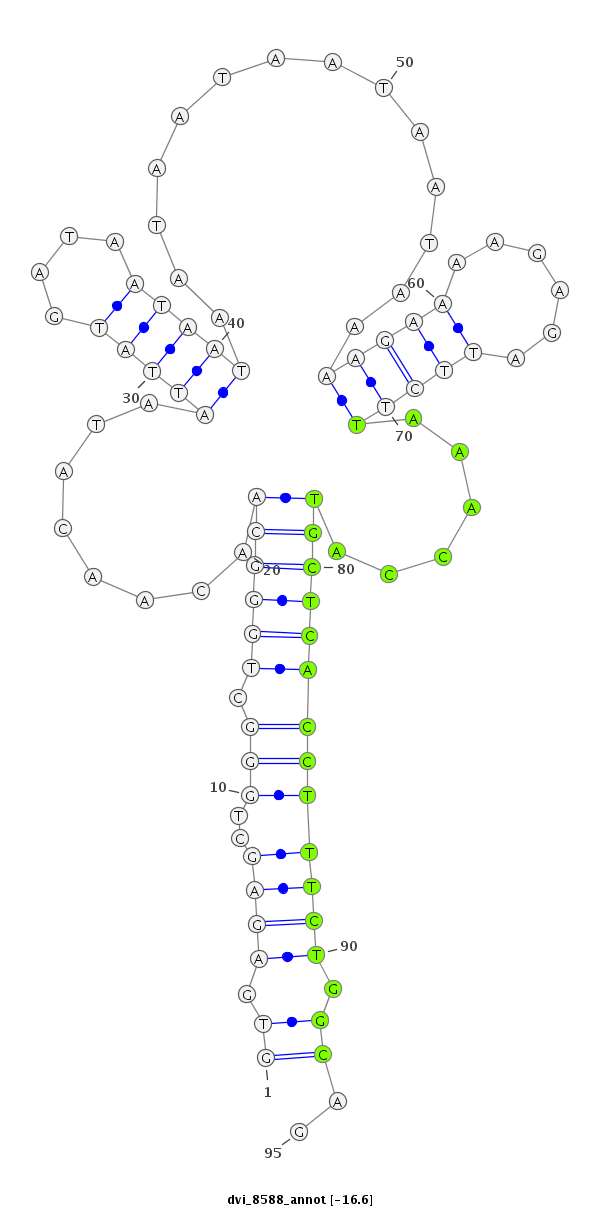 |
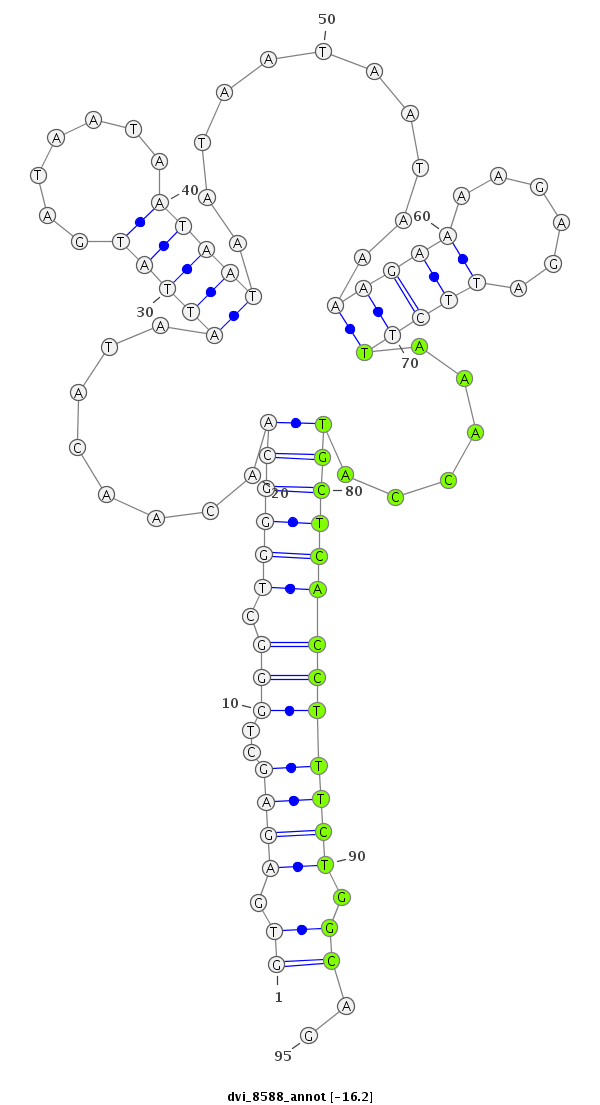 |
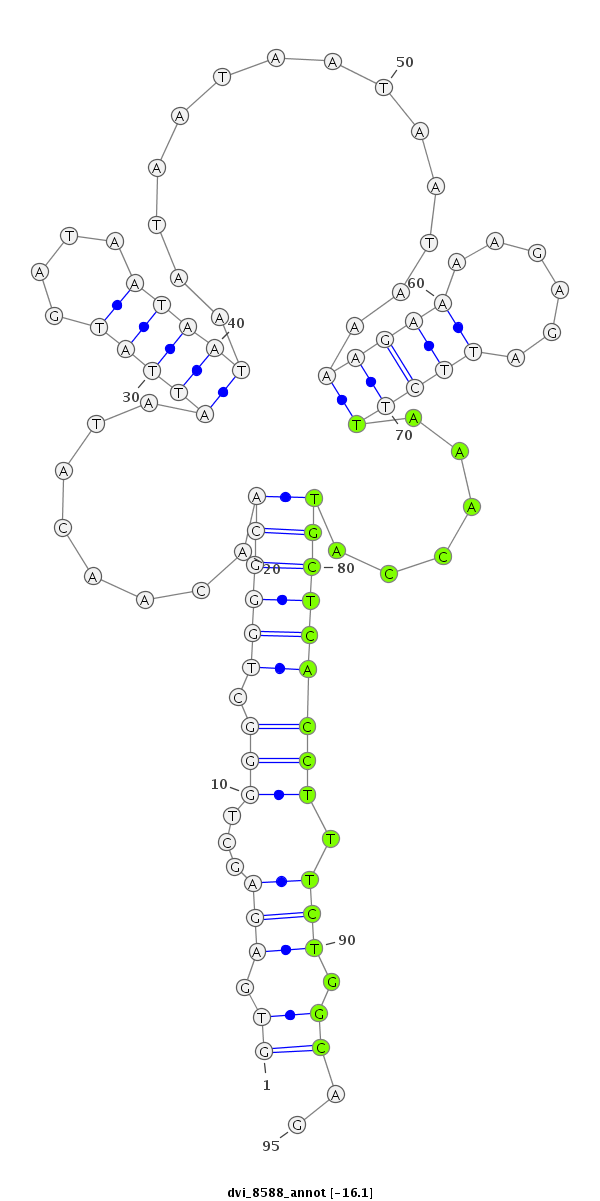 |
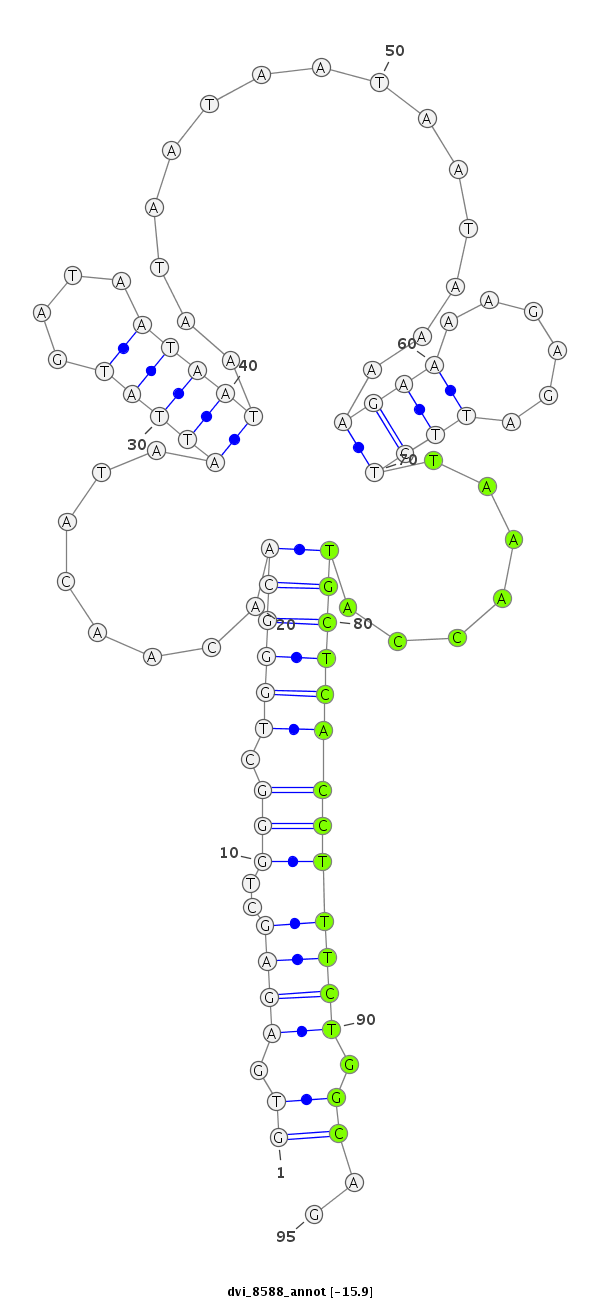 |
exon [dvir_GLEANR_17314:2]; CDS [Dvir\GJ16809-cds]; CDS [Dvir\GJ16809-cds]; exon [dvir_GLEANR_17314:1]; intron [Dvir\GJ16809-in]
| Name | Class | Family | Strand |
| (TAA)n | Simple_repeat | Simple_repeat | + |
| ##################################################-----------------------------------------------------------------------------------------------################################################## TGGCCATGGTGCTGGGCTATCAGCTGCAAGAGGAGCACTTGCACTCGTCGGTGAGAGCTGGGCTGGGCAACAACATAATTATGATAATAATAATAATAATAATAAAAGAAAAGAGATTCTTAAACCATGCTCACCTTTCTGGCAGTTTGTGTGCAGCATCTGCAAGGTGAGCCTGCTCAACTATGAGGAAATGGC **************************************************.((..((..(((.((((((..........................................................)))))))))..))..)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
SRR1106719 embryo_8-10h |
V047 embryo |
V053 head |
SRR060677 Argx9_ovaries_total |
M047 female body |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TAAACCATGCTCACCTTTCTGGC.................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCTATCAGCTGCAAGAGGAGCACTTG.......................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................GGTGAACCTGCTCAAGTAT........... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................ATATGCAAGGTGAGCATTCTC................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GTGTGTAGAATCTGCAATGTG.......................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................GCTCCCCGTTCTGGCTGTTT............................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GGAGGGCTTGGCTGGGCAAC............................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................CTCAGCTATGTGGAACTGGC | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................AACAACAGAATTATGATACG........................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
ACCGGTACCACGACCCGATAGTCGACGTTCTCCTCGTGAACGTGAGCAGCCACTCTCGACCCGACCCGTTGTTGTATTAATACTATTATTATTATTATTATTATTTTCTTTTCTCTAAGAATTTGGTACGAGTGGAAAGACCGTCAAACACACGTCGTAGACGTTCCACTCGGACGAGTTGATACTCCTTTACCG
**************************************************.((..((..(((.((((((..........................................................)))))))))..))..)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................GAATTTGGTATTAGTGGAAA......................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAGAATTTGGTACGAGTG............................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................GACGAGTTGATACTCCTTTAC.. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............CCGATAGTCGACGTTCTCCTC................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................GATAGTCGACGTTCTCCTC................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................AGCGGCCGCTCTCGACCCC.................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................TTAATACTATCATTCTTA..................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:2126267-2126461 + | dvi_8588 | TGGCCATGGTGCTGGGCTATCAGCTGCAAGAGGAGCACTTGCACTCGTCGGTGAGAGCTG--------------GGCTGGGCAACAACATAATTATGATAATAATAATAATAATAATAAAAGAAAAGAGATTCTTAAACCATG---CTCACCTTTCTGGCAGTTTGTGTGCAGCATCTGCAAGGTGAGCCTGCTCAACTATGAGGAAATGGC |
| droMoj3 | scaffold_6473:2006538-2006701 + | TGGCAATAGTCCTGGGCTATGAACTGCAGGAAGAGCAGCTTCACTCCACGGTGAGCATAAGAGAGGAAGAGTGTTCCTTAATATCAATG--------------AACTCA-------------------------------ATA---CTCTCATATTTCGCAGTTTGTGTGCAGCACCTGCAAGGTGAGCCTGCTCAACTTCGAGGCATTAGA | |
| droGri2 | scaffold_15203:8533914-8534065 - | TAAGCCAGATTTTGGGCCACAAACTGCACGGAGAGCAATTGCATTCCACGGTGAGTCCTG--------------ACCTGAATTTCAGCG--------------AATCGA-------------------------------ACTTGAACCCACTTTGTTGCAGTATTTATGCAGCGGCTGCAAGATGGACTTGCTTCAATATGAAACA-TATC | |
| droBip1 | scf7180000395187:41996-42046 + | TATTTTT-----------------------------------------------------------------------------------------TATTAAAAAAAGGAAAATGATAAAAAAAAAGAGATTGGTGAACC------------------------------------------------------------------------ | |
| droEle1 | scf7180000491268:1115572-1115605 + | AATATAA-------------------------------------------------------------------------------------TAATAATAATAATAATAATAATAATAA--------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/16/2015 at 08:09 PM