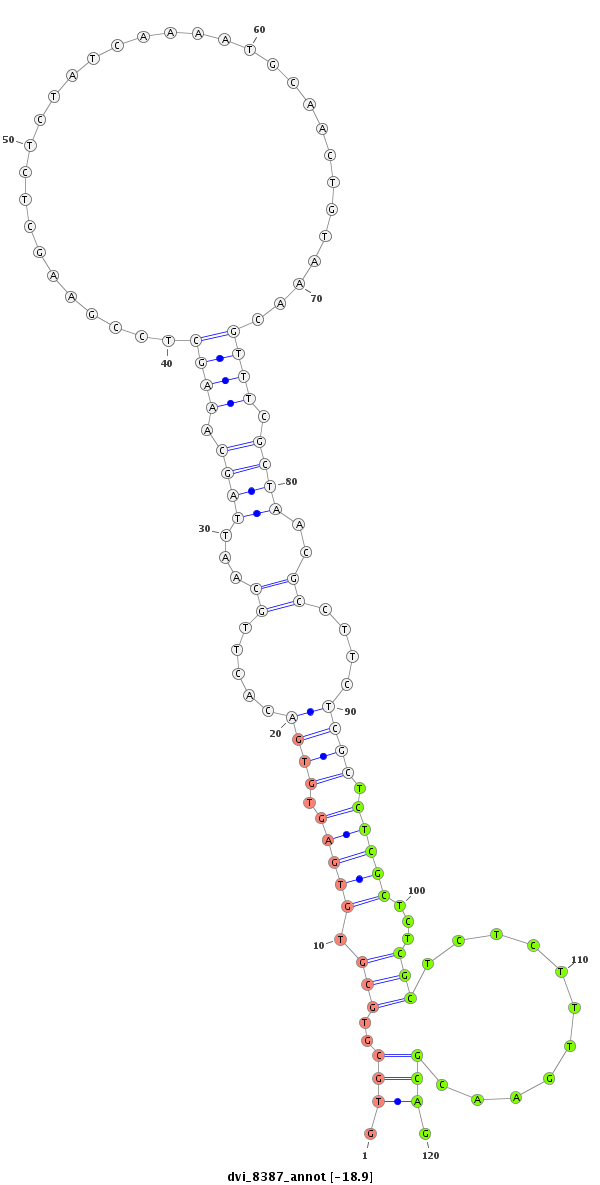
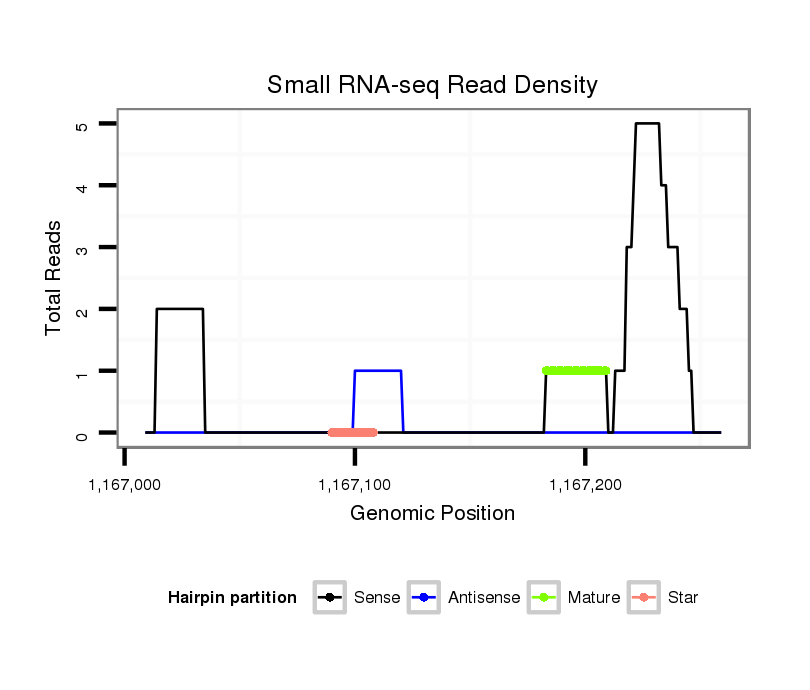
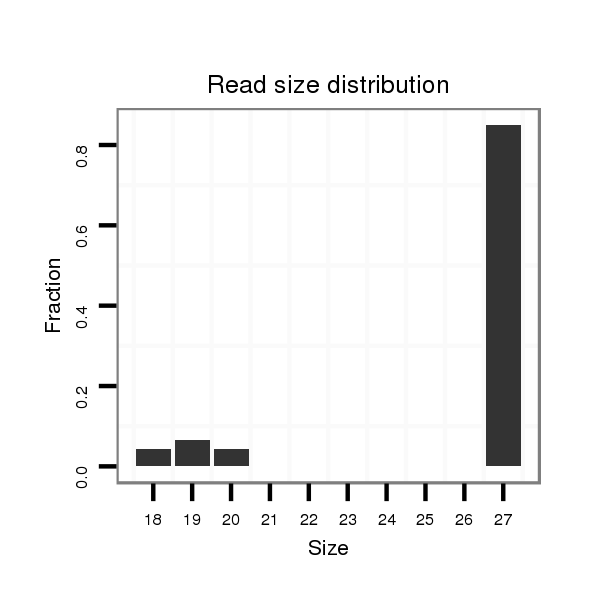
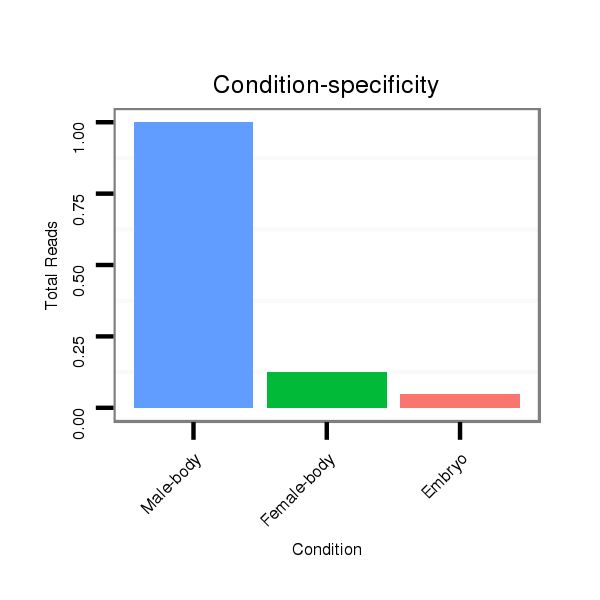
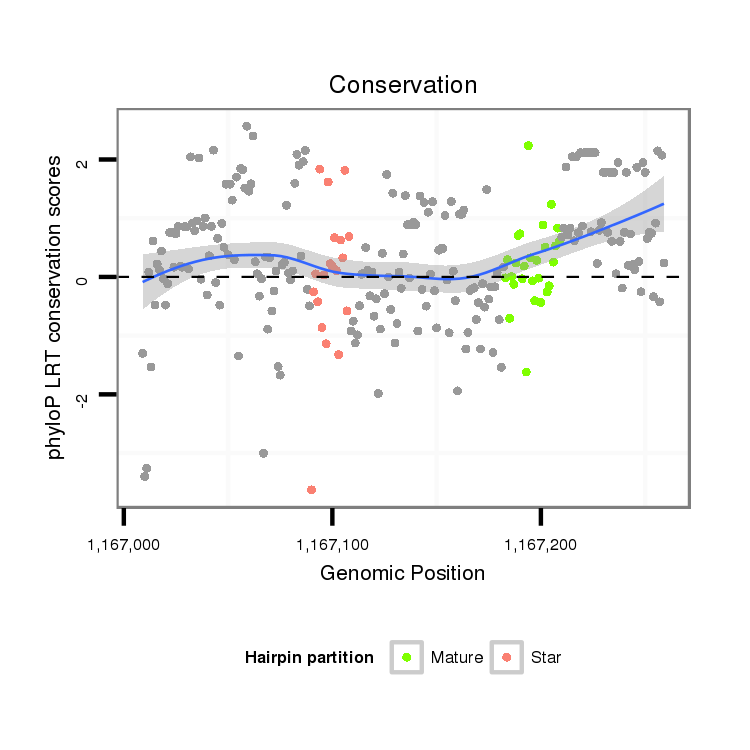
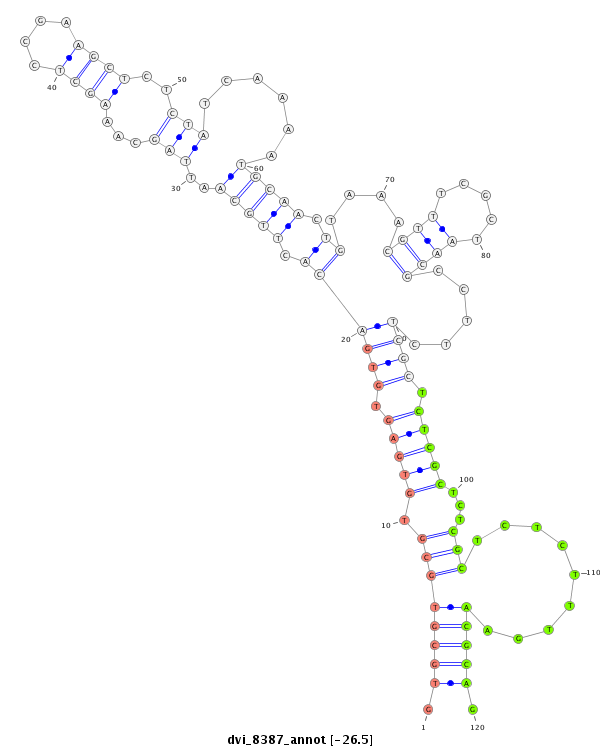
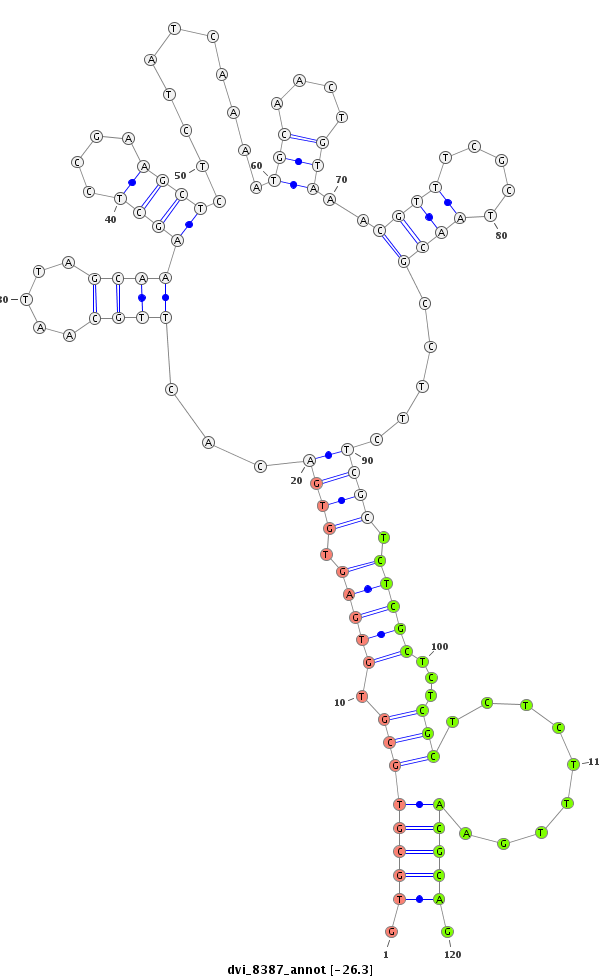
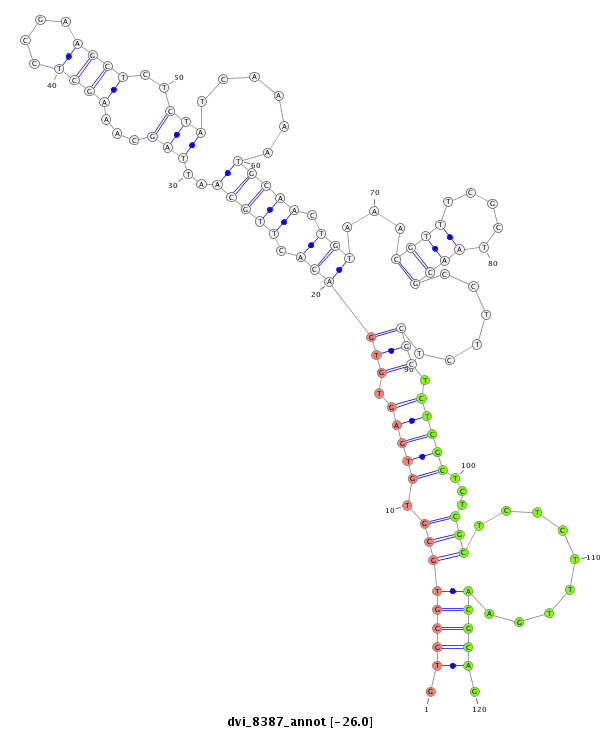
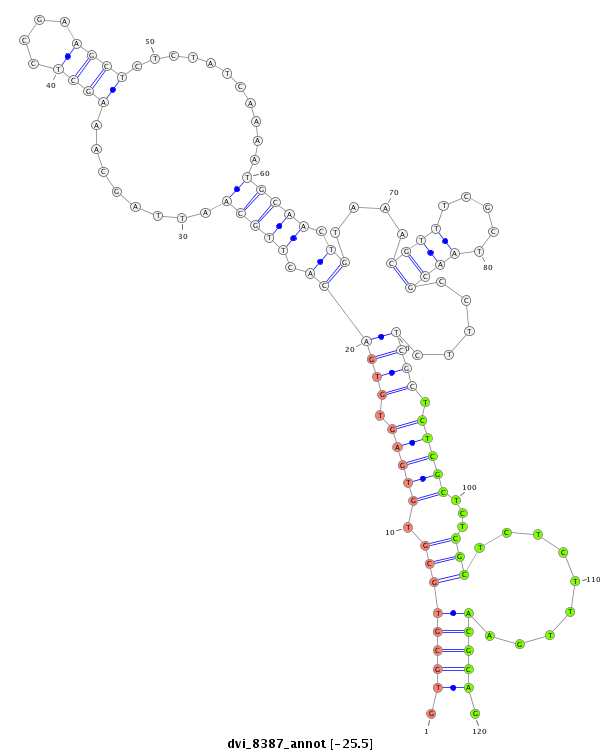
ID:dvi_8387 |
Coordinate:scaffold_12928:1167059-1167209 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.5 | -26.3 | -26.0 | -25.5 |
 |
 |
 |
 |
exon [dvir_GLEANR_17276:3]; CDS [Dvir\GJ16768-cds]; intron [Dvir\GJ16768-in]
| Name | Class | Family | Strand |
| (TG)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAACAATTTTGACCGCCAGCTTGTTGAGCGTAAGTGTGTGTGTGTGTGTGTGTGTGTGCGTGCGAGTGCGTGTGTGTGTGCGTGCGTGCGTGTGAGTGTGACACTTGCAATTAGCAAAGCTCCGAAGCTCTCTATCAAAATGCAACTGTAAACGTTTCGCTAACGCCTTCTCGCTCTCGCTCTCGCTCTCTTTGAACGCAGGTGGATCTGATATTCAACGAGGAGGGCTCCTGCCCGCTGTGCAACAAGAC *********************************************************************************.(((..(((.(((((.((((.....((...((((.((((.................................)))).))))..))....)))).)))))...)))...........))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060670 9_testes_total |
V053 head |
M047 female body |
M027 male body |
V116 male body |
SRR060658 140_ovaries_total |
SRR060663 160_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060656 9x160_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060665 9_females_carcasses_total |
V047 embryo |
SRR060662 9x160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....ATTTTGACCGCCAGCTTGTTG................................................................................................................................................................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GACCGCCAGCTTGTTGAGCGTGG.......................................................................................................................................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GATATTCAACGAGGAGGGCTCCT................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GATCTGATATTCAACGAGGA........................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................ATTCAACGAGGAGGGCTCCTGCCC............... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TCTCGCTCTCGCTCTCTTTGAACGCAG.................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GATATTCAACGAGGAGGG........................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................TTCAACGAGGAGGGCTCCTGCCCGC............. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTTGTGAGTGTGACACTGGCT.............................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGCGTGTGACACTGG................................................................................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTCAGTGTTACACCTGC............................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................GAAGCACTCTACCAAGATGC............................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................TGTATGTGTGTGCGTGCGAGTGC...................................................................................................................................................................................... | 23 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGAGTGTCACAGTTGCT.............................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGAGCGTGACAGTTG................................................................................................................................................ | 19 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTCTGAGTGTTACACTTGCT.............................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGAGTGTGACAGCTGCCA............................................................................................................................................. | 22 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAGCTCCCAAGTTCTCCATC................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................CTTGTGAGTGTGACACCCGC............................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGAGTGTGACGGTTGCT.............................................................................................................................................. | 21 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTTGTGAGTGTGACAATTCC............................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................GTGCGTGCGTGTGAGTGTG....................................................................................................................................................... | 19 | 0 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................TCTTTGAACGCACCAGGAT............................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........ACAGCCAGCTTGATGAGCTT............................................................................................................................................................................................................................ | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGTGTGTGCGTGCGAGTGCA..................................................................................................................................................................................... | 20 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCACTGCCAGCTTGATGAGC.............................................................................................................................................................................................................................. | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................TCAATAAGGAGGGCTCATG.................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTGTGCGTGTGACAGCTGCA.............................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GTGTGTGTGCGTGCGTGC.................................................................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................CGTGTGCGTGTGACAGTTGCT.............................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TATCACAGCCAGCTTGTTG................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TCGCTCTCGCTCTCGCTCTCTA........................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TGTGTGTGCGTGCGTGCGTG............................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................GTGTGTGTGTGTGCGTGCG........................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGTGTGTGTGTGCGTGCGAG......................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTTGTTAAAACTGGCGGTCGAACAACTCGCATTCACACACACACACACACACACACACGCACGCTCACGCACACACACACGCACGCACGCACACTCACACTGTGAACGTTAATCGTTTCGAGGCTTCGAGAGATAGTTTTACGTTGACATTTGCAAAGCGATTGCGGAAGAGCGAGAGCGAGAGCGAGAGAAACTTGCGTCCACCTAGACTATAAGTTGCTCCTCCCGAGGACGGGCGACACGTTGTTCTG
**************************************************.(((..(((.(((((.((((.....((...((((.((((.................................)))).))))..))....)))).)))))...)))...........))).********************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
V116 male body |
SRR060677 Argx9_ovaries_total |
M061 embryo |
SRR060675 140x9_ovaries_total |
SRR060654 160x9_ovaries_total |
SRR060679 140x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................CGGGCGAGACGTTGTTCAG | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CACTCACACTGTGAACGTTAA........................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................ATCGCGTAAGAGCGAGAGCGTGAGCG................................................................. | 26 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AGAGCGAGAGCGAGAGCGAGAGCAA.......................................................... | 25 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................ACGCTCACGCACACACACAC........................................................................................................................................................................... | 20 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................GCTCACGCACACACACACGCTC....................................................................................................................................................................... | 22 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................AGCGAGAGCGAGAGCGAGAGA............................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:1167009-1167259 + | dvi_8387 | CAACAATT------TTG-----ACCGCCAGCTTGTTGAGC--GTAAGTGTGTGTGTGTGTG--------------------------------------TG------------------T--GTGT----------GTGCGT------GCGAGTGCGTGTG--TGTGTGC--G--TGCGTGCGTGTG------------AGTGTGA----------------------------------------------------------------CACTTGCAATTA-GCAAAGCTCCGAA--------------G-------CTCTCTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAA--------------------AATG-----------------------CAACTGTAAA-----------CGTT-TCGCTAA--------------------CGCCTTCTCGCT--C---------------------------------------------------------------------------------------------------------------------------TCGCT----C--T------------------C----GCTCTC-----T--T-T--G------AACGCAGGTGGATCTGATATTCAACGAGGAGGGCTCCTGCCCGCTGTGCAACAAGAC |
| droMoj3 | scaffold_6473:13920494-13920734 - | CTGCCATT------TTG-----ACCGACAGCTTGTTGAGC--GTAAGTT----------AGT--T---C-AG---------CCTCTCTGGCTACATGTGTGCTT--------------GTGT--GTGCC------------------------------TG--TGTGTCC--C--TGTGCGTGAGTG------------AGTGTGA----------------------------------------------------------------CACTTGCAATTA-GCAGAGCTTCAAA--------------G-------CTCTCAA-------------------------------C-----------------------------------------------ACC----A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATT-----------------------CAACACTAAC----------------------------------------------CGTCTCGCT--T---------------------------------------------------------------------------------------------------------------------------CCGCT----C--T------------------C----GCTCTC-----A--T-T--T------GACACAGGTGGATCTGATATTCAACGAGGAGGGCTCATGCCCGCTGTGCAACAAAAC | |
| droGri2 | scaffold_14853:8378351-8378598 + | CAACCATT------TTG-----ACCGCCAGCTTGTTGAGC--GTAAGTTTCT------------AA---T----------GC--------------------GTGTGCG--------TGTGT--GTGTT------T------GTGTAT--A--------------------------------------------------TGCGG----------------------------------------------------------------TACTTGCAATTA-GCAAAGC----------------------GCTCTT--------------------------------------T-TGCTCTCTCTCTCTCTCTCGTCTCTCTCTTTCTCTCTTACCAAAACAAA----AAATGCATTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAAT---------------------------------------------------TGTCAACA--------------CTCGCG-----------------------------------------------------------------------------------------------------------CGCTC--TC----------------------TC--------------------ATTCAACACAGGTGGATCTGATATTCAACGAGGAGGGCTCCTGCCCGCTCTGCAATAAGAC | |
| droWil2 | scf2_1100000004909:4247486-4247808 - | CGACGACC------GCC-----AGCGCCAGCTTGTTGAGC--GTGAGTTTCT--------------------------------------------------GTGTGTG--------TGTGT--GTGTG------T------GTGCA--TA------------TGTGTTT--T-----GTA--TG--------------------AT---------------------------------------------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCG--------------------AGAAAGACAGAG------AG-------AGAGAGAGACAGTGAGAGTAAGCTCTCTAGCAATGCTAAACAAGGCTTACTCTAACTCCCTGC---TCTCTCTCTGTTCCTA--------------------GACCCCACCCCACCTCCCACCC---------------------------------------ACC----------------TACCTACCTACC--C---------------------------------------------------------------------------------------------------------------------------CCGCC----G--CCCCACTGTAACATCATTAT----TCAAACTATATATTCTCCTGCTTTACACCACAGGTGGATCTGATATTCAACGAGGAGGGCTCTTGCCCACTGTGCAACAAAAC | |
| dp5 | XL_group1a:7222917-7223172 + | CCATATTT------TTG-----GCCGCCAGCTTGTTGAGC--GTGAGTT-CTG----------------------------TGTGT----------------TT--------------GTGT--GTGT--------GAGAG--------------------------------------AG----------GGAGTGCA--------------------------------------ACAGTGT------------------------------------TT-GCCAAAATTTACA--------------GTTATAAC--------------------------------------T-CCCCCACCCTCACTCAATCGTACTCCTGTTCCCCTCCCACGCAATCACTTGCTCACTGCGTATGTG---C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCTTTTTTTTCT--------------CTTTCT-----------------------------------------------------------------------------------------------------------CGCTC--TT----------------------GC--------------------CCGCGTCACAGGTGGATCTGATATTCAATGAGGAGGGGTCCTGCCCCCTTTGCAATAAGAC | |
| droPer2 | scaffold_13:1834416-1834679 + | CCATATTT------TTG-----GCCGCCAGCTTGTTGAGC--GTGAGTT-CT----------------------------GTGTGT----------------TT--------------GTGT--GGGTGTG----T------GTGTGT--GAG--------------------------AG----------GGAGTGCA--------------------------------------ACAGTGT------------------------------------TT-GCAAAAATTTACA--------------GTTATAAC--------------------------------------TCCCCCCACCCTCACTCCATCGTACCCGTGTTCCCCTCCCACGCAATCACTTGCTCACTGCTTATGTG---C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTCTTTT-TTTCT--------------CTTTCT-----------------------------------------------------------------------------------------------------------CGCTC--TT----------------------GC--------------------CCGCGTCACAGGTGGATCTGATATTCAATGAGGAGGGGTCCTGTCCCCTTTGCAATAAGAC | |
| droAna3 | scaffold_13337:8530466-8530660 - | TGT---GT------G----------------------------------TGTGTGT----------------------------------------------GTGTGTG----------T--GTGTGTG------T------GTGAG------------TG--TGTGTTC--T--CTCGTAGGTGTG------------AGTGCGTATCTCAAAGA--TACGAAAAGTCCAATACAAATAGTTTTTAAAAAAAGATACGTGAAGGTGCTTCCGTTTCACTCTCCT------CT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCGCTCTCTCT--C---------------------------------------------------------------------------------------------------------------------------TCTCT----C--T------------------C----TCTCTC-----T--C-T--G-----------TCTCGAGTGTGACATTTAACAA------------------------------ | |
| droBip1 | scf7180000396566:234400-234505 - | TAA---GT------G----------------------------------GGCATGTGTGCG--------------------------------------TG------------------T--GTGT----------CTAGAT------GCGATTGTGCCTG--TGTGTGC--G--AGTGTGGGTATG------------AGTGTGT----------------------------------------------------------------GTCTCGCG---------AGTTTTT-----------------------------CA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GACATTTAAT--------------------------TGAAAA | |
| droKik1 | scf7180000302696:884277-884419 + | GCG----------------------------------------------TATGTGTGTGTG--------------------------------------TGTGTATGTG----------T--GTGTGTG------T------GTGTA------------TG--TGTGTGC--A-------GTGTGCA------------AATGCAA----------------------------------------------------------------TGTTTGCACTCAGCT------CGAGA--------------G-------CACTCAA-------------------------------T-----------------------------------------------TAG----A-------------------------------------------------------------------------------AA---------------------------------------------------------------------------------------------------------------------------------------------CGTT-AAGCTAA--------------------CG----------------------ACGACC--------------------------------------------------------------------------------------------------GCTGCTC---TCACT----CTTC------------------C----TCTCAC-----T--C-T------------------------------------------------------------------ | |
| droFic1 | scf7180000453323:125026-125267 + | TTC---TT------TTG-----GCCGCCAGCTTGTTGAGC--GTAAGT----------GGGAGGGGAATA----------GTGATA----------------TT--------------GTGT--GG----------------------TTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTAAGCTATT-------CTAGTGCCAGATTTGAATACATTCG---TGTCAAGTTTAAAATGTCTTATAAATAACGA--AAAACTGGAGTAAATAA-------AAAAACAAA---------------------------------------------------------------------------------------------------------------------------CAGTAAA-------------------CAAA--------------------AC----------------------ATCTA--------------------------------------------------------------------------------------------------CTTTGTAC---CTAT------------------TCTTC---------------------------------------AAGGTGGATCTGATATTCAACGAGGAGGGCTCCTGCCCGCTATGCAACAAAAC | |
| droEle1 | scf7180000490996:895266-895472 + | TAGCTAAG------TTGACGTCGCCGCTGCACTCTTGTGT--GTCAGT----------GTGT----------------------------------------GTGTGTG----------T--GTGTGTG------T------GTGTGTGTA--------------AGTGA--T--TGTGTA----TGAGTG--------CGAGTG------------------------------------CGT------------------------------------TT-A----------------------------------------AATTGAAAATTCTGCTAGGCGAAATGGGAATATCCCCCTTCCCTTTCCAATCCCCCCATACATTCCACCCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCTAAACCCCT--C---------------------------------------------------------------------------------------------------------------------------TCGCT----C--C------------------C----TCCCCT-----T--T-T--G---------------------------------------------ACTGCA------------ | |
| droRho1 | scf7180000779914:481247-481466 + | AACCGAATTGAAAGTTTATGTCA---------------GC--GAAAA--------------------AT-GGCGGCTCTA-TCTGTCTGTG--------TGTGTGTGT----------------GTGTG----------TGT------GTGACGTTG------TGTGTGTATTAGTACGAATGTGTG------------AGTGCGCAACCCGACGTCA-------------------AATATGT------------------------------------TT-GAACAACTTTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATGCAA-------AATGCCAAA---------------------------------------------------------------------------------------------------------------------------CATTCTCCAACTTGTAGACGTG-GCGCTCT--------------------CGCCTCTCTT-TCCCC--------------CTTTCT------------------------------------------------------------------------------------------------------------------C------------------C----GCTTTC-----T--C-T--G--------------------------------------------------------------- | |
| droBia1 | scf7180000298389:351406-351580 + | TGT--------------------------------TGAGT--GTGTGT----------GTGT-----------------A-TCTGTGAGAGTGTGAGAGTGTGT--------------------------------------GTGTGT--GAG------AGTGTGTGTG------TGTGCGTGTGTG------------TGTGTGTA---------------------------------------------------TCTGTG---TAATGGCAGGATGCAACA------TGAAAATGGCGTGTGGCTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAACGTTT---------------------------TATT----------------------------------TGCTCGTTCTCTGGTACT--------------------------------------------------------------------------------------------------TTT-------------------C------------------CCTGCGCTCCT-----T--T-T--G--------------------------------------------------------------A | |
| droTak1 | scf7180000415704:939764-939881 - | GATTGATT------TAA-----ATTGGCAAGTACTAAAGT--ATGTGTGTGTGTGT----------------------------------------------GTGTGTG----------T--GTGTGTG------T------GTG----TGTGTGTGTGTG--TGTGTGT--G--TGTG--------------------GGA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGGGGCGGTGGAGCTTGCGATGTGCTAA--- | |
| droEug1 | scf7180000409089:124732-124880 + | CGC---AT------GTG----------------------------------TGTTTGAGTG--------------------------------------T--GTGTGTGTGTGTGTGTGTGT--GTGTGTGTGTGT------GTGTGT-------------------------------AG----------GGGGTGTG--------------------------------------GCAGCG-------------------------------------TA-GCAAAGG----------------------GTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------CCCCCCTCATAGACATCTAATGAGTGTTGG-----------------------------------------------------------------------------------------------------------------------------------G-TCCTTTA--------------------CA----------------------GTAACC--------------------------------------------------------------------------------------------------TCTCCTC---TC----------------------------------TCTCTC-----T--C-T------------------------------------------------------------------ | |
| dm3 | chrX:4806519-4806825 + | TTC---TT------TTG-----GCCGCCAGCTTGTTGAGC--GTGAGT----------GGGAGTGGGA------------------------------------------------------------------------GG------GCGAT--------------------------------------------------------------------------------------AGTGA--------------------------CACA------------------------------------GT---------------------------G-----------------------------------ATGTGCACTTGGCTACT-------TATTCGTTAGAACTATATGCATATATAATGTACAGCTT--------------------AGAA-------AAAAGG--TTTTTCGCATAAAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATAGTAGGTTGTATTAAAGTAAAATGGCATATC----AT------ATATAATGCCTTTCTGATGGAAGGTAACCCGCGGTAGAAGGCAAGCCCTTTGCAACTAATGTT----CGAT----TTGTTTCTTT---------------------------------------AAGGTGGATCTGATATTCAACGAGGAGGGCTCCTGCCCGCTATGCAACAAAAC | |
| droSim2 | 3r:6225109-6225290 + | GTG----------------------------------------------TGTGTGTGTGTG--------------------------------------TG------------------T--GTGC----------GAAGGG------GCGACTGTGTGCG--TGGGTGC--T--TGTGTGAGCATA------------AGTGTGG----------------------------------------------------------------CACT------CTGCC------C-----------------------------------------------------------------------ACG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTGCCTGCACTTCTAGCTCTGCGCCGA--------------------GAACGAGCCGAGCCCCCCGCCCCAATT---------------------------CGAC---CCCCAGCTGGCTGCCCCAGCCCCTTCTTCCC--T---------------------------------------------------------------------------------------------------------------------------TCGCT----C--T-------------------------------------------------------------------------------------------------------CAC | |
| droSec2 | scaffold_0:20611118-20611305 - | GTG----------------------------------------------TGTGTGTGTGTG--TTG---T----------GT------------------------------------------GTGTG----------CGT------GTGCTCG-----------------C----------------------------TGTGTT---------------------------------------------------TGTGCG---GAACATCAACTAATATTG------TCAACGCAACATTCGACTTGTTATCCC--------------------------------------TTCCGCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-GCCTGACTTC-----------------------CTCT---TCTTAATTTCGTT-----------------------------------TTGAACTCCTCGACGTCAGCGATTTTGTC----------------GGCCTTCTCGCT--T---------------------------------------------------------------------------------------------------------------------------TCGCT----C--T------------------C----TCTCTC-----T--C-T------------------------------------------------------------------ | |
| droYak3 | X:3638838-3639157 - | TTC---TT------TTG-----GCCGCCAGCTTGTTGAGC--GTGAGT----------GGGAGTGGGA------------------------------------------------------------------------GG------GCGAT--------------------------------------------------------------------------------------AGTGC--------------------------CCCA------------------------------------GT---------------------------G-----------------------------------ATGTCCACTTGGCTACTGGCTACCTAATCACCAGATCTATATGTATATG---TGTAGTGCTT--------------------AGAAGGACAGGAAAAAGTATT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCT--------------TTTTTTTGATAAGTTCT-TAAAAGTACTATGGCATATCTACCATATATGCATATAAAACCTTTTGAATGGGAGA--GCC--TAGCAAGAGGCAAGCACTTT-CAACTGATGCT----CTAT----TTGTAACTTC---------------------------------------AAGGTGGATCTAATATTCAACGAGGAGGGCTCCTGCCCGCTATGCAACAAAAC | |
| droEre2 | scaffold_4929:8551182-8551373 + | GCT---GA------TTG-----ATCA-----------AGTGTGTGGGT----------GTG--------------------------------------T--GCGTGTG----------T--GTGTGTG------T------GTGTG------------TG--TGTGTGT--C--CGTGTGCGTGTG------------CGTGTGTC---------------------------------------------------TGGCTG---GGGCACAAACAAAA-----------------------------------------------------------------------------------------------------------------------------------ACCATA--T----------------------------------------------------G--TTTCT--------------------------------------------------C-GCCCGACTCC-----------------------CGCACGCACTCACTCACAAA-----------------------------------CCGAATTACTC----------ACTCAGCC----------------CGCCTCCTC---------------------------------------------------------------------------------------------------------------------------------TCGCTCACTC--T------------------T----GCTTTC-----G--T-T--G--------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 03:39 AM