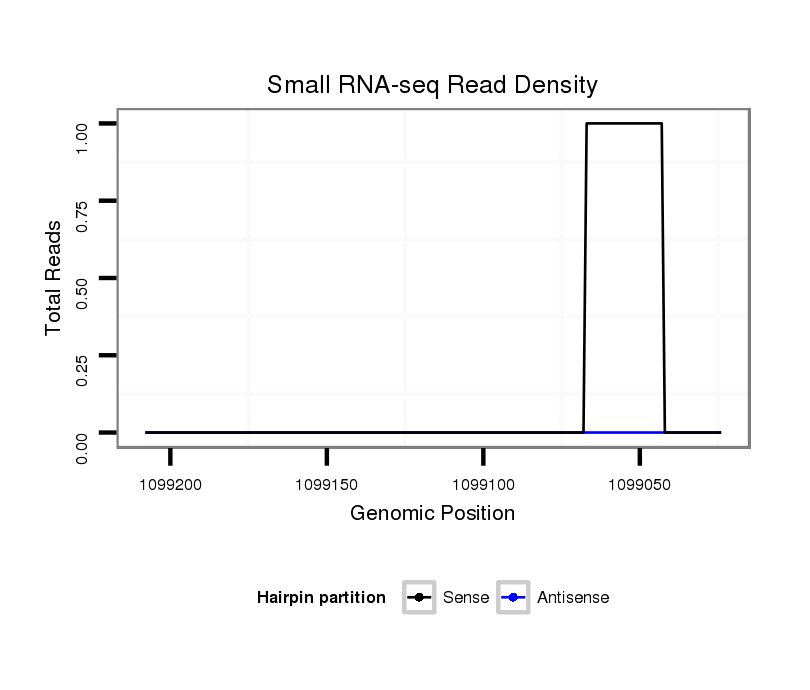
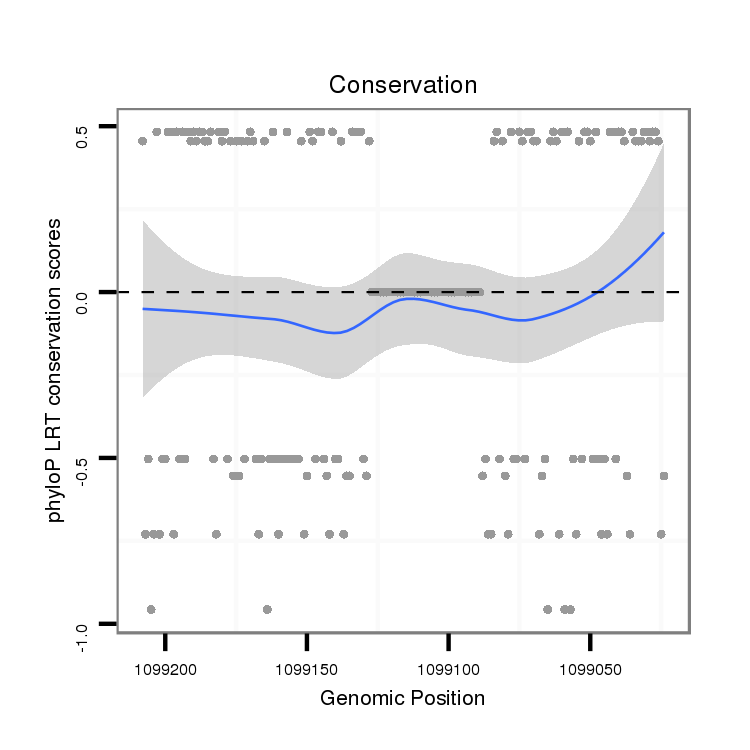
ID:dvi_8379 |
Coordinate:scaffold_12928:1099074-1099158 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ16652-cds]; exon [dvir_GLEANR_17156:3]; CDS [Dvir\GJ16652-cds]; exon [dvir_GLEANR_17156:4]; intron [Dvir\GJ16652-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------################################################## CCACAACCATACTCTATCAGATCCACATGAACTGTCGCTGGCTCGGACAGGTTGGCCCTTCGATTTAATGCCATTATATTCCATAACCGTAATAATATATGCATTTGATTTGATATCGTTTTCGCTGCTGTCCAGATTGGCTCCCAGGTCAGCGCTATGGTGAGCATATTGAATCGCATCAAGCT **************************************************...(((....((((..(((((...((((((.............)))))).))))).........)))).....))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
V116 male body |
V053 head |
V047 embryo |
SRR1106728 larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................CTGGCTCGGAGAGGTTGGTA................................................................................................................................ | 20 | 3 | 2 | 4.50 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| .....................TTCACATGAACTGGCGCTG................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TCCCAGGTCAGCGCTATGGTGAGCA................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................CATAATATTCCATAACCTTGAT............................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................TGGCTCAGAGGGCAGCGCTA............................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................CGGTGGGTCGGATAGGTTGG.................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................CGATCCACATGAACT........................................................................................................................................................ | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................CGGGTCATCGCTATGGTGG...................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GGTGTTGGTATGAGATAGTCTAGGTGTACTTGACAGCGACCGAGCCTGTCCAACCGGGAAGCTAAATTACGGTAATATAAGGTATTGGCATTATTATATACGTAAACTAAACTATAGCAAAAGCGACGACAGGTCTAACCGAGGGTCCAGTCGCGATACCACTCGTATAACTTAGCGTAGTTCGA
**************************************************...(((....((((..(((((...((((((.............)))))).))))).........)))).....))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M027 male body |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................AGTCGCGCTACCACTCTTAC................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 |
| ....................................................................ACGGGAATAGATGGTATTGG................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 |
| ................................................................................................................................ACAGGTCTTGCCGTGGGTC...................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:1099024-1099208 - | dvi_8379 | CCACAACCATACTCTATCAGATCCACA------------------------TGAACTGTCGCTGGCTCGGACAGGTTGGCCCTTCGATTTAATGCCATTATATTCCATAACCGTAATAATATATGCATTTGATTTGATATCGTTTTCGCTGCTGTCCAGATTGGCTCCCAGGTCAGCGCTATGGTGAGCATATTGAATCGCATCAAGCT |
| droMoj3 | scaffold_6328:2078183-2078352 - | CAGGCAATGTAATTTGTCAGATCCATCCCGATGACCATTGGACTAATCTGGTGAGCAGACACTGAACCCAATCAATCAATCAATCAATCACACACATATATACAC---------------------------------------ACATCTACATTTTAGGTTGGAATGCAGTTGACTTCCATGATACAAATGTTGTCTCGCATCAAGAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 11:54 PM