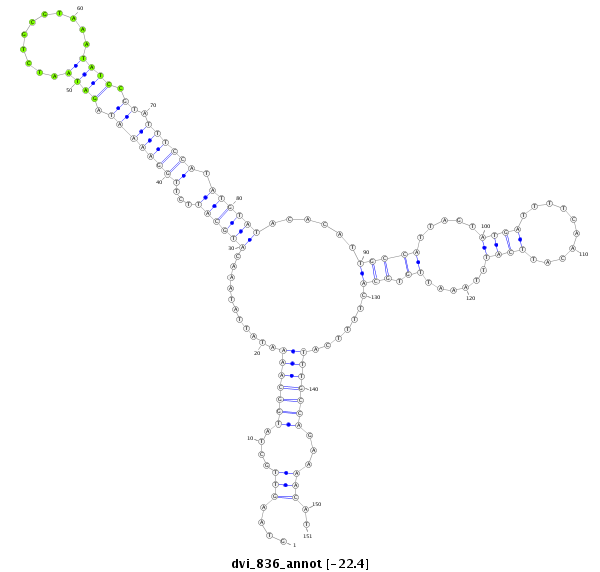
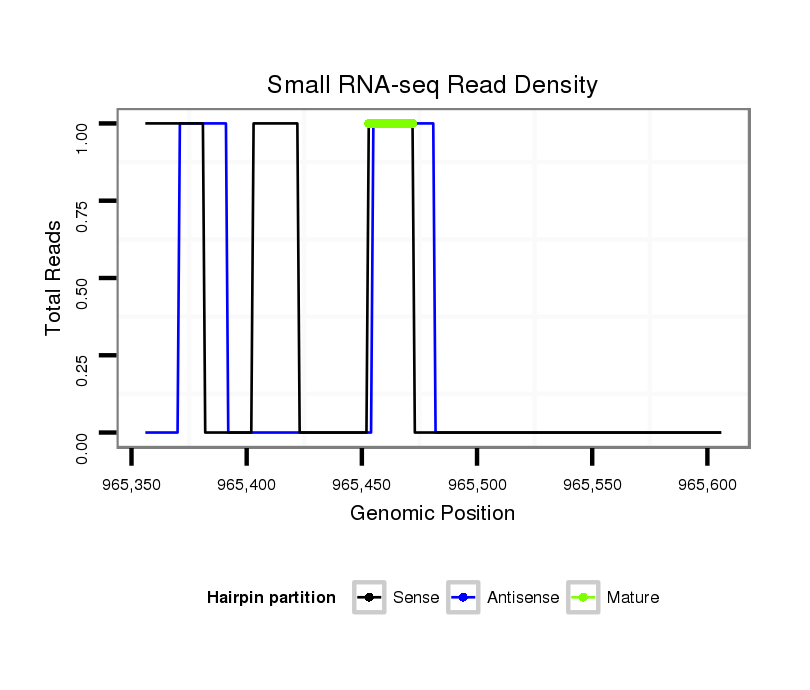

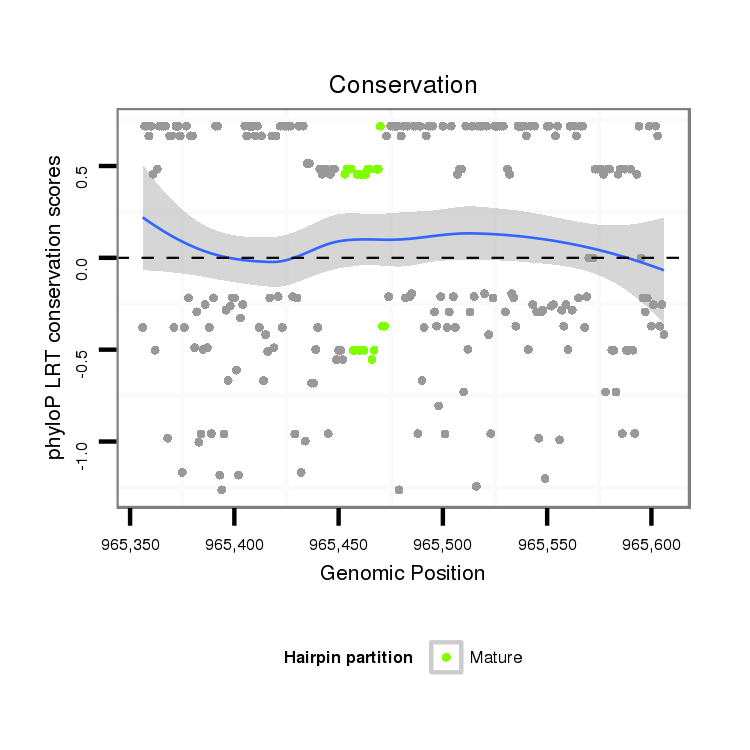
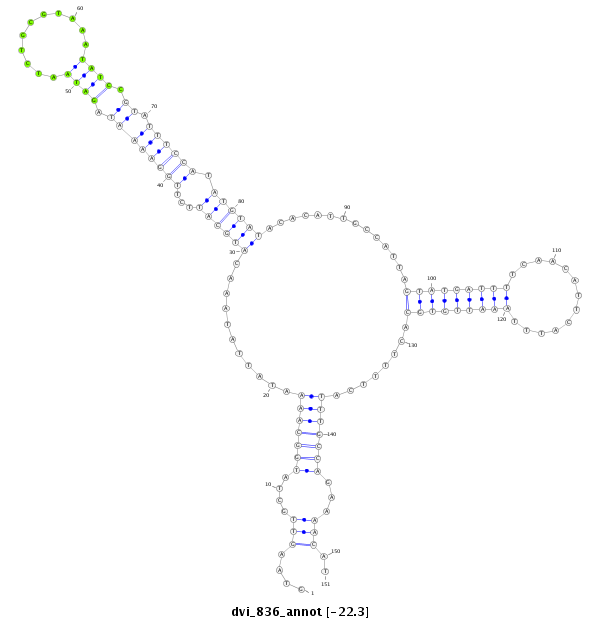
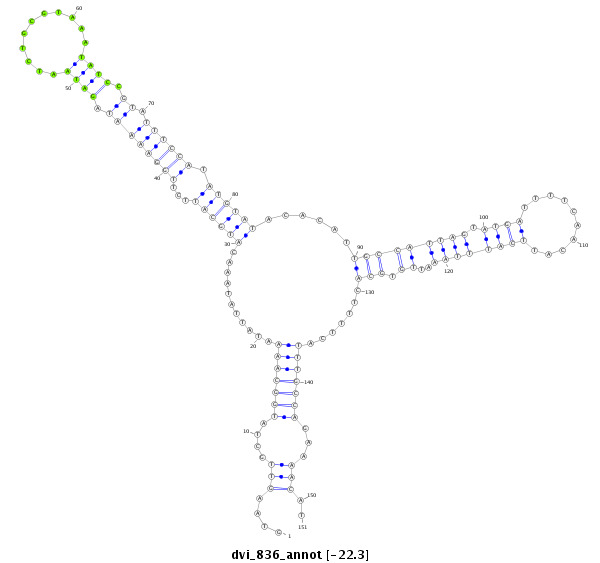
ID:dvi_836 |
Coordinate:scaffold_10324:965406-965556 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.3 | -22.3 | -22.1 |
 |
 |
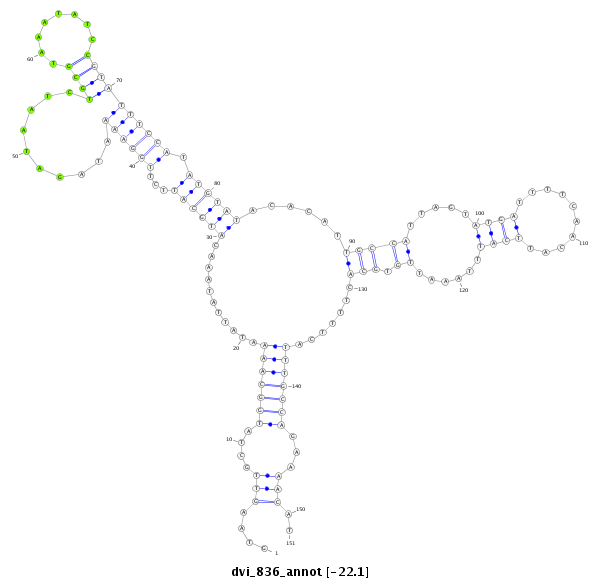 |
CDS [Dvir\GJ15114-cds]; exon [dvir_GLEANR_15413:1]; intron [Dvir\GJ15114-in]
No Repeatable elements found
| ----------------##################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TATGTCGTATAACCGTATGAAAACGCCAAAAGATTTAACAACTGTCACAAGTAAGTTGCTATGGCAAAATATTATAAACATGCATTCTTGGAAAATAGATAATCTGCGTAAATATCCGTATTTCCATATGTATACACATTGCCATTAGTATGATTTTCAACATTCATTTAAATTGTGCACTTTTCATTTGCCAGAAAACATTCAAAATGCATTTAACATTTGAAATCAGAGTTCTGCCAGTGTACATGCAA **************************************************....(((....(((((((...........((((((...((((((((.((((...........)))).)).)))))).))))))......(((((.....((((..........))))......)).))).......)))))))...)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
M047 female body |
SRR060678 9x140_testes_total |
SRR060680 9xArg_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TATGTCGTATAACCGTATGAAAACGC................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CAAGTAAGTTGCTATGGCAA........................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........ATAACCGTATGAAAACGCCCAAAG........................................................................................................................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........ATAACCGTATGAAAACGCCCA.............................................................................................................................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................GATAATCTGCGTAAATATCC...................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................TGCGTTAATATCCGGATTT................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................ACCATTTGAAATCAGTGTT.................. | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAAATAGATAAGTTGCGC.............................................................................................................................................. | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................ACCTATGGAATCAGAGTT.................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATACAGCATATTGGCATACTTTTGCGGTTTTCTAAATTGTTGACAGTGTTCATTCAACGATACCGTTTTATAATATTTGTACGTAAGAACCTTTTATCTATTAGACGCATTTATAGGCATAAAGGTATACATATGTGTAACGGTAATCATACTAAAAGTTGTAAGTAAATTTAACACGTGAAAAGTAAACGGTCTTTTGTAAGTTTTACGTAAATTGTAAACTTTAGTCTCAAGACGGTCACATGTACGTT
**************************************************....(((....(((((((...........((((((...((((((((.((((...........)))).)).)))))).))))))......(((((.....((((..........))))......)).))).......)))))))...)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
SRR060676 9xArg_ovaries_total |
V053 head |
SRR060673 9_ovaries_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..ACAGCATATTGGCCTACTTTTGCGGGT.............................................................................................................................................................................................................................. | 27 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............ATACTTTTGCGGTTTTCTAAA....................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................ATTAGACGCATTTATAGGCATAAAGGT............................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..ACAGCATATTGGCATACTTTTGCGGGT.............................................................................................................................................................................................................................. | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................GACTCAAGACGGTCACCGGTA.... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................AACGGACTTTTGTAAGGTTT............................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................CTACGATACCGTTTTGTTATA................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................CCTAAACTGTTGACAGAGTT......................................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_10324:965356-965606 + | dvi_836 | TATGTCGTATAACCGTATGAAAACGCCAAAAGATTTAACAACTGTCACAAGTAAGTTGCTATGG---CAAAATATTATAAACATGCATTCTTGGAAAATAGATAATCTGC-GTAAATATCCGT----ATTTCC--------------ATA----TGTATACACATTGCCATTAGTAT-------------------------------------------GATTTTCAACATTCATT--TAA-----------------------------------------ATTGT--GCACTTTTCATTTGCC-----------------AGAAAACATTCAAA-------ATG------------------CATTTAACATTTGAAATCAGAGTTCTGCCAGTGTACATGCAA |
| droMoj3 | scaffold_6496:807436-807811 + | TATGTCATATAATCGTATGGAAACGCCCTCGGACTTACGTGAAATGCAGAGTAAGTTGAA-TGGATTCAAAATATTTTAGAA----CTTCTTCGAATGCTGATAGCCCGTAGTATGTATTTGTGTATATTTGCATATACATAC----ATATGTGTGTACAAACATTGTGATAAGTATTGTGGCATTTCAAATGCTGTGAGGCATATTCATATAGAATATTGATGTGCAAAATTCAACCATAAATTGCTGATGGAGCATAAGCTCATAAACGTAGATGTGCATAATTGTCGGCATTTTTCCTCTGTCCCGTTTTAACTACAGGGGAAATGCAACTGCATACGTGTGTG------------------TATTT---ATTTGCAACTCGACTCTTAGCA-TGTATGTGTGT | |
| droGri2 | scaffold_15245:6101025-6101260 + | GATGT---ATAAACGGATGCCATCGGATGAACCAATAGAGGATGACGCAAGTAAGTGGAATAGC---CGACATATCGAACATATTAAG------------------------------TTTGCACATATTTACCAACACACCCCCCAAAATGTACATAAAACCATTTTGGTCCTTGGC----------------------------------------------TTAAGGATTTAAA--AAA-----------------------------------------ATTT----TTTTTTTCATTTTAA-----------------GCAAAACAGCTAAATAAATAAGTGAGCCGAATGATAGAAATGCATGC------------------------A-ATAATATGTAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 10:21 PM