| droVir3 |
scaffold_12928:965598-965848 - |
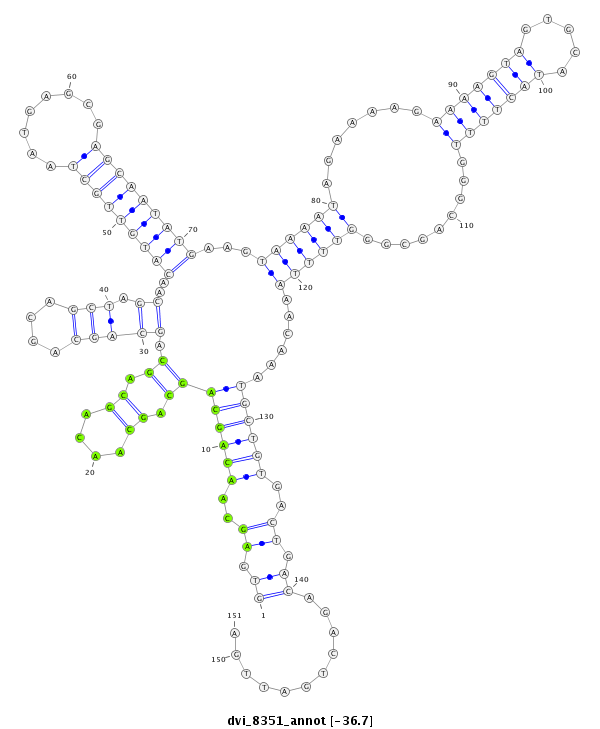
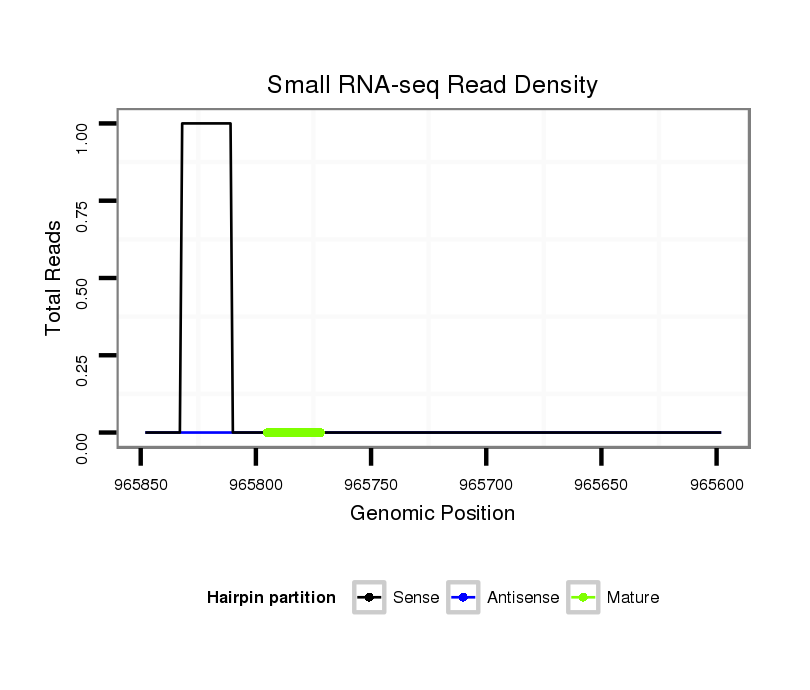
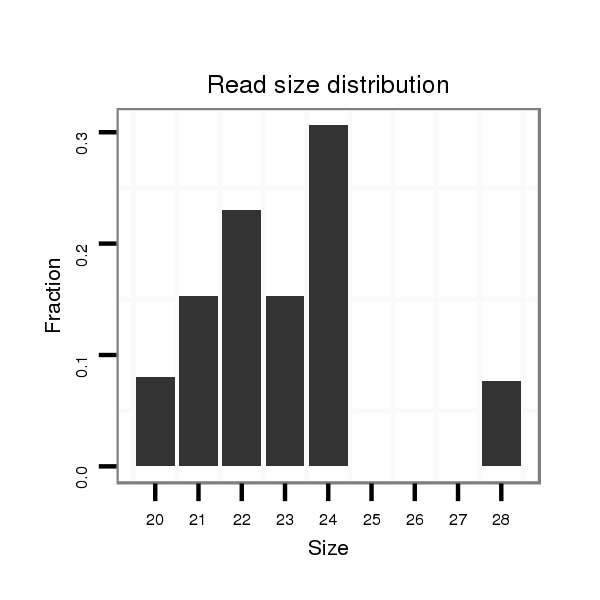
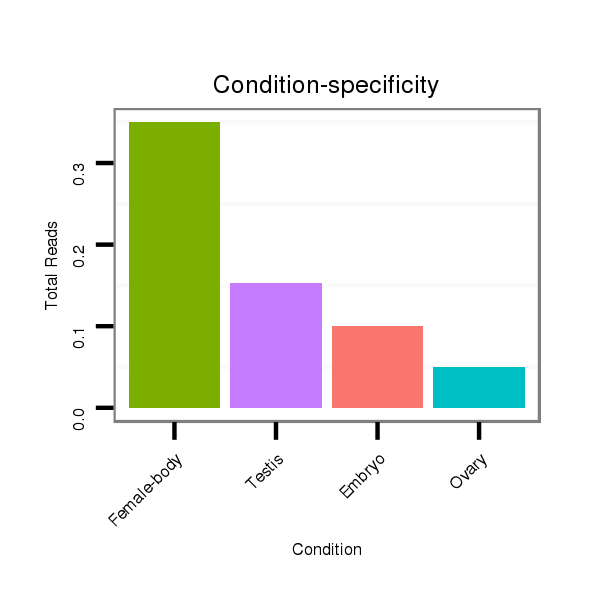
dvi_8351 |
CAAT------------------------AACGGTA----ATCACA--------A----------------------G-----A--AGGACA--AGCGCAGCAGCC--AT-TAT---AATTCAGGTGAG----------------------------CAACAGC---AGCAGCAACAGCAGCAGCAGCAG---CA-------------------------------------------------------------------------------GCT-------AGCAACATG--------------TTGCTAATGAGCGAGCAATATGAAG------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATAGAAAAGAAAAGTA---------------------------------GTGCATACT---------------------------------------------TTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGCAGC------------------------GGGTTTTAAACAAATGCTGT-----------------------------------------------------------------------------------------------------------GA----CTGACAGACTGATTGACTGACTGAC--------TGACT--------AA----TTGAACAATGCTAGTT----GTCTAAAAAACTGAAATG |
| droMoj3 |
scaffold_6540:16946520-16946780 + |
|
CAGT------------------------AGCGACG----GCAGCA--------G----------------------C-----A----------------------GCAGGAGCA-G---AA---------G---CA----------------GCAGCAGTAGC---GACAGCAGCAGCAGTAG---CGA---------------------------------------------------------------------CAGCAGCAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGGAGCAG-------------------------------------------------------------------------------------------------------------------CAGC--A-------------------------------------------------------------------------------------------------------------------------------------------------------GGAG--------------CA---------GCAGCAGTAATACAGCGCATCTTGACTGACGACGTCGACTGGG--------GCGCGCTCAATGGCTCACTAAACTTTTGA--------------------------------------ATAAAAGCAGG-CATGCAGT---------------------------------------------------------------C----GCTTGTCGG------------------GCGGGCTGGCTG------------GCTGGCTGGCTGGCTGGCTGGCT--------GGCTGGCTGG------CTGGCT----GGCTGGCTG-------TC |
| droGri2 |
scaffold_14853:9295862-9296087 - |
|
CGTTGTGTTGTTATTTGATTGTTGCCAGAGCAG-------------------------------------------------------------------T--------------------------------------------------------AGC---AGCAGCAGC------AGCAACG---GCAGCAGCAGC---------------------------------AGCAGCAGTAGCAGCA---GCAGC------------------------AGCAGCAAC------------------------------------ATGAAG------------C----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAGGAAA---------------------------------------------------------------------------AGGAAAACG----------------------------------------------------------------------TCGTCCAAATGG----------G------GAAATGAGAAAAA--------------------------------------CTGCACACATTGGCT---------------------------------------------------------------------GGTTGTTTGTCACGCTGCTCTGTCACA-------------------------------------------------------------------------TGACTGACTG----ACTGACTGACTGACTGA----------------------------CTGA------TTAAC--------------------TGA |
| droWil2 |
scf2_1100000004514:1993977-1994172 + |
|
AGC-------------------------AACATC------------------------------------------------------------------------------------------------------------------------AGAAGCAAC---AACAGCAA------CAG---------------------------------------------------------------CA---GC---------------------------AGCAACAGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAT------------------------------------ATC----AAAC---------------------------------CAACAAGCAACAGAAAAAAGG---------------------------------------------------------------------------------------------------------------------------------------------------------AAGCAATTTCA-----------------------------------------------------------------------------------------------------------------------GGCT--AAAT-----------------------------------------------TTAGTTTAAATGCCTACCCGTTCATTCCAGCCATTGGCCGTTTGCCGG------------------AGAGA------------CTGACTGACTGACTGAC--------TGACTCGCTGGATGA----CTCG------TTGAC--------TGACG-----ACTTG |
| dp5 |
XR_group8:3709124-3709285 - |
|
CAGC------------------------AGCAGCA----GCAGCA--------G----------------------C-----A-------------------GAC--AGCGGCCGC---CA---------C---TA----------------GCTCCAACAAC---AACAGCAACAGCAACAA---CAA---------------------------------------------------------------------CAACAGCAATCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC---------------------------------AGACAAGCAAC---------------------------------------------------------------------------------------------------------------------------------------------------------GGGAA--------------CA---------ACAACGAGACAAC-------------------------------------------------GGTT-CCTGCACCTTTGGGCAGCAGCGGCC------------------------ATGCTAGTGCTGC-----------------------------------------------------------------------------------------------------------CA----ATG-------------------------------------------------------------------------------------GC |
| droPer2 |
scaffold_21:1288447-1288634 - |
|
CCAA------------------------AACCAGA----TGAAGA--------T------------------------------------------------GAA--GACGACAAA---AA---------G---GA----------------GCCACAACAGC---GGCAGCAGCAGCAGCAG---CAA---------------------------------------------------------------------CAGCAGCAACAAGCTACC-------TGCAACATG--------------T----------------------TG------------C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGTGCA-----------------------------------------------------------------------------------------------------------------------GGTA--AAAA-----------------------------------------------T-------GTTG------------------CCTTTTGTTATTTGCCGACATCCAACCACAACTGGCTCTGG----CTGGCTGGCTGGCTGGCTGT--------------------------------CTGG------CT----------------GG------CT |
| droAna3 |
scaffold_12929:911680-911822 + |
|
CCAT------------------------AACGGAGAACAATC-CA--------C----------------------G-----A--CCCACA--CACACAAAGGTC--ATCTTCAAT---CC---------G---TA----------------TCCCTACCAGC---AACAGCAACAGCATCAGCA---TCAGCA------------------------------------------------------------------CCAGCAACATGTCCAGGAACAGGAGCAAC------------------------------------ATGTCG------------T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAATTTGGAGGAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------AA |
| droBip1 |
scf7180000396589:2060506-2060674 - |
|
CAAC------------------------AACAGCA----ACAACT--------T----------------------G-----T--GGG-----AGCGCAGCA---GCAACAGCA----------------------ACAGATCCTG-------------------------------------------------------------ATTCAGCAGCAACAA---CCACA---------------GCA---GCAGCAACAGCA---------------------------GATATTGCAGCAAAAGTTGCCC-------------------------------------------------------------------------------------------ATCGATCCGCAACAGCAGCAAC---------------------AACA------------AC---------------------------------------------------------------------------------------------------------A-----------------------------------------GCAGCAGCAGCAAA-----------------------TC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGCAAC------------------------AGGTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C--TAA |
| droKik1 |
scf7180000302686:1552489-1552734 + |
|
CAGG------------------------CAG-ACA----GCCAGACTGGCGACGGTGGCATATTCCCGTGGGGAGCTGCATTTAA-------------------------------AAGGCATTTAAGCCACAAAA----------------TCTCAAGCAGC---AACAGCAA------------CAA---CA---------GC--------CAGCAACAA------------------------------------TTACAGCAAG-------------------------------------------------TAGGGGAGATTAATTCAAGCTGATCGG---AAAAGCTGGAATACTCTAAAAAAGATTTTTAAAAGATTTCTGTAAAATTTAAACTCTTTAATTCACAAAAAAAAAACTATAA---------------------------------A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAAAGAAGAAATTA |
| droFic1 |
scf7180000453940:65473-65645 + |
|
AGAA------------------------A----------------------------------------------------------------------------------------------------------------------------CTACGGGAACAGCGGCAGC------AGCGACAACAG---------------------------------------------------------------------CTGCAAAAATGAGCTGAC-------GGAAGCGAG--------------C---------------GATACGATG------------C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGCCACGAGAAG------TGAATG---------------------TCTTCTGGGGGA----------------------TTGACTGACTG----GCTGACTAACTGACTGAC--------TGACT--------AA----CTGAGTAA--CTGAGT----AACTGACTGACTGAATTA |
| droEle1 |
scf7180000491193:71263-71515 + |
|
CAAC------------------------AACAAGA----GTTACA--------T----------------------T-----TAA-------------------------------AAAAAAAGAAAAACACA--------------------------------------------------------------------------------------------------------------------------------------------------CACTCACAT--------------------------------------------------------------------------------------------------------------------TCA---CCGAAAAAAA------ACAACAACAATAAT----------CACGCGCGGCAA--CGCGGCACGCAGCACCTGCTTTAAAAAGAAGTGCGAGTGGGGGGAGCGAGGTTAAAAAAGTATTGAAAACTGGAGAAAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT------------------------------CC------------------------AG-----------------------------------------------------------------------------------------TTGTCTGATTGATAC------------------GATGA----CTGACTGACTAACTGACTGACTGAT--------TGACT--------AA----CTCA------CTGACT----GTCTAACTGACTGACTGA |
| droRho1 |
scf7180000780178:26582-26730 - |
|
CAAC------------------------ATCAGCA----ACAGCA--------G----------------------C-----A---------------------------------------------------------------------------------------TC------AGCAACA---GCAGCATCAGC---------------------------------AACAGCA------------GA-----------------ATC----AACAGCAACAACAGG------------------------------------TTA------------TGCA--------------------------------------------------------ATTG---TACAATAATA------ACAACAACAACAACAGTAACAA-------------------------------------------------------------------------AA---------------------------------AGAA---------------------------------GCG-----------------------------------------------------------------------------------------------------------------------------------------------CAAGCGTAACAACA-----------------------------------------------------------------------------------------------------------A-----AAACAAAAGCTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GA |
| droBia1 |
scf7180000302408:2042625-2042792 - |
|
CGTT------------------------GACAGTG----AACACG--------G----------------------A-----G--GGGAC-CAGGAGCAGCA---G-----------------------------------------------------------------------------------CAGCAGCAGCAACACCCAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCAACAGCA------GCAGCAACAGCAAC----------TGC--------------------CAC-----------------------------------------------------------------------------------------------------------------------------CTCCTG--CCTTGAAGG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C----------------------ATGACAAACTG----ACTGACTGACTGACTGAC--------TGACT--------GA----CTGA------GTGACTGATGGCCCGACTGTCTG--CAA |
| droTak1 |
scf7180000415660:122960-123081 - |
|
CAAT------------------------GGCGTCT-----CCGCA--------G----------------------C-----A-------------------GAC--AACTTTT----------------------ACGGA---------------CAGCAGC---AGCAGCAACAGCAGCAG---------------------------------------------------------------CA---GA-----------------GCTACC-------AGCAACATG--------------T---------------AAGTAGAAC------------A-------------------------------------------------------------------------------TTAAAA---------------------------------A---------------------------------------------------------------------------------------------------------CAAGAAAGG---------AAGCT--------ACC---------------------------------------------TTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GC |
| droEug1 |
scf7180000409466:3091974-3092162 + |
|
TGAT------------------------AACAACA----ATCAGC-------------------------------TACAGCTAC-------------------------------CAGCCAAGTGGACCATA-------------------------------------------------------------------------------------------------------AAA------------AA-----------------GTA----ACCAGCAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCGGCAGAGAGGC----TCTTAAAAAGCAGCAGTAGCAGCGGGCAAAAGCTGCGAAACGGGCGGAAAAGAAAAGGAAAATTTCAAATGAAGCGCA---------------------------------TTGCATTTA---------------------------------------------TCC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTCTC------------------------GAGCTCTCAACGA--------------------------------------------------------------------------------------------------------GA----------------------------------------------------------------------------------------------TG------CT |
| dm3 |
chrX:10512415-10512584 - |
|
CAAC------------------------ACTAGCA----ATCACA--------A----------------------G-----A----------------------ACAACAGCA-A---AC---------G---CTGC-------------AGT---CGC---AGCAGCAGC------AGC------------------AGCAG------------------CAGCTGCA---------------GTAGCAGC---------------------------AGCAGCAGC-------------------------------------------------------------------------------------------------------------------------------------------------------------AG-------------------------------------------------------------------------------------------------------------------CAGC--A---------------------AC---------GCTAG--CGTCGGCAGCAGCAT--------------------------------------------------------------------------------GTCCAAAACAAGTAGCGGTAATCCAGCA--------------------------------------------------------------------------------------------------GCAGC------------------------AGGTG--CAACAAATGCCGC-----------------------------------------------------------------------------------------------------------GA-------------------------------------------------------------------------------------------ATG |
| droSim2 |
3r:3574966-3575162 - |
|
CAAT------------------------CACAATC----ATCACA--------T----------------------G-----C--------CAAGAGCGGTA---GCATCAGCG----------------------GCAGAATTTCAAA-------------------------------------------------------------------------------------------------------------------------GCC----ATCGGCCACAT--------------------------------------------------------------------------------------------------------------------CCG---CCCAACAGCA------ACAACAGCAGCAAC----------AGC--------------------AAC------------------------------------AGC----AAAT---------------------------------GCGTAAGAAAGACAAAGATAGATCGAATAGTAGTT--------GC---------ATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCAGTGGTAGTCATGTTGGTGCTGGTAGCGGTAGTCAAGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATAG--TGT |
| droSec2 |
scaffold_13:1772649-1772898 - |
|
ATGT------------------------TGCTGCG----GTGGCT--------G----------------------G-----T--GG-------------------CACTTGTT----------------------GCGGCTTGCTGCGGTTGC------------CGCTA------------------------------CTG------------------CTACTGCAACTGCAACTGCAGCGGCA---GCGGC------------------------AGCAGCAGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTC---------------------------------------T--------------------------------------------------------------------------------------GCGTG--TCAATTAGC-G---TTGATGATGCACAGTGAAATAGATTTCGCGC--A-GCTTGTTTCAATTCCAATTCGTTCGACAATTGTTACAGCTAAAGATCTACAAAACAT-CAGCGCCTATCAAGTGGAAGTGTCA-----------------------------------------------------------------------------------------------------------------------AGTG--T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAACAAAACAAAAACG |
| droYak3 |
2R:15026131-15026354 + |
|
CACT------------------------CTCCGCC----GCCCCC--------T----------------------C-----G--G--------------GA---ACAACAGCA----------------------ACATGTTGTTAGGTCAGC------------CGCAG------------------CAGCAGCAGCAACAA---------------------------------CAAGAGCAACTGCAAC---------------------------AGCAGCAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------ACGA------------AC---------------------------------------------------------------------------------------------------------A---------------------TC---------TCTAGGCCATTGGCAGTG---TG-----------------------GC--ATGCAA----------------------------------------------ATGGGAGCTGTGCAGCGTAGTTCCAACGGC-------------------------------------------------TTGGAGCAGGGAGCTGGAT-------------TCAGTCTATCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGAATAATTCTGCTT----GTTGGAGAAACTG--TAG |
| droEre2 |
scaffold_4690:4615484-4615636 + |
|
GCAG------------------------T--------------------------------------------------------------------------------------------------------------------------------CGC---AGCAGCAGC------AGC------------------AGCAG------------------CAGCTGCAGTAGCAGCAGCAGCAGCA---GCAGC------------------------AGCAGCAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTAG--CGTCGGCAGCAGCAT--------------------------------------------------------------------------------GTCCAAAACAAGCAGCGGTAATCCAGCAGCAGCAGCA-----------------------------------------------------------------------------------------------------------------------GGTG--CAACAAATGCCGCGAATGCCGC---------------TGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT |