
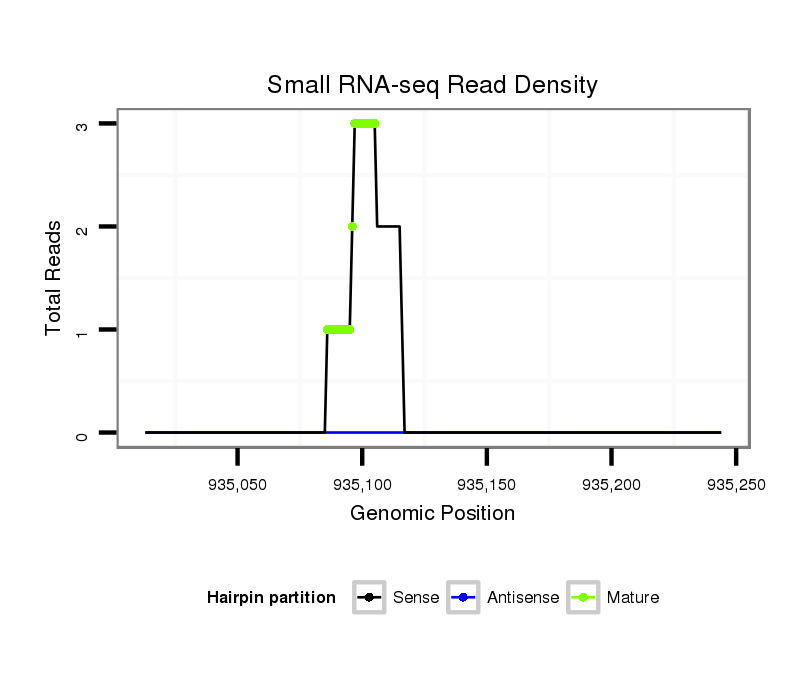
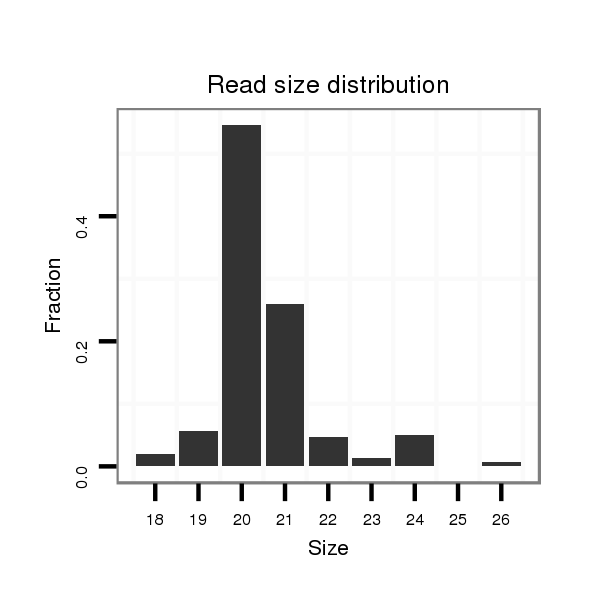
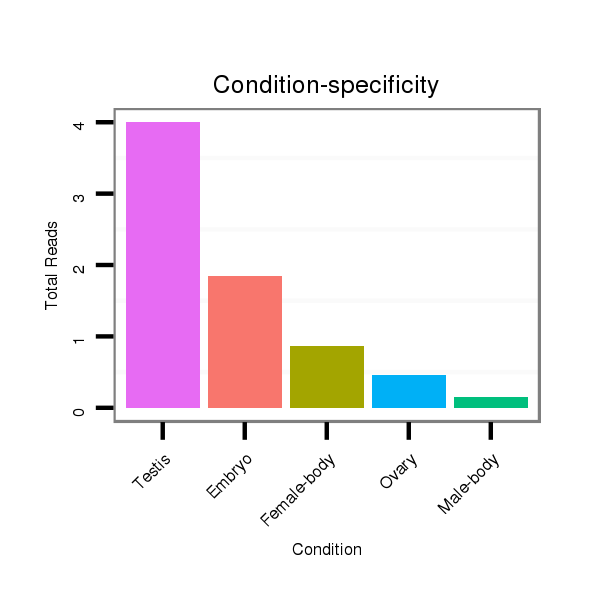
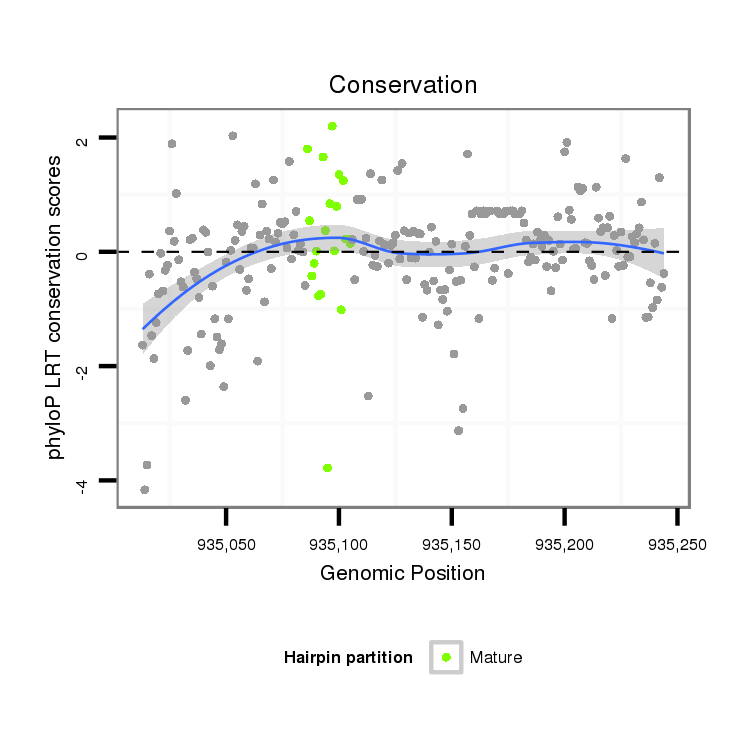
ID:dvi_826 |
Coordinate:scaffold_10324:935063-935194 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.5 | -28.0 | -27.8 |
 |
 |
 |
CDS [Dvir\GJ15109-cds]; CDS [Dvir\GJ15109-cds]; exon [dvir_GLEANR_15409:1]; exon [dvir_GLEANR_15409:2]; intron [Dvir\GJ15109-in]
| Name | Class | Family | Strand |
| (ATG)n | Simple_repeat | Simple_repeat | + |
| ##################################################------------------------------------------------------------------------------------------------------------------------------------################################################## ATTGCCTGCTCTATGCTGCCCGCATAATTGCCAATGACAATGATGATGATGTTGCTGATGATGATGTTGCTGATGATGATGATGATGATGATGAGGGGAATGCGTATGTGAGTGCTGGCCATGACCATGACGTCAATACACAAATACAAACACATTTACGCAATACGCAAGCCGGCAACAAGCACGACAACATTGACGCTAATGACGAGGTGTCAACGGCACGTGAACGACA **************************************************((((((.............((.((.((.((....)).)).)).)).((..((((((((((((((.((....((((......))))...............)))))))))...)))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060659 Argentina_testes_total |
SRR060681 Argx9_testes_total |
M061 embryo |
SRR060685 9xArg_0-2h_embryos_total |
V053 head |
M047 female body |
SRR060667 160_females_carcasses_total |
SRR060655 9x160_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060666 160_males_carcasses_total |
SRR060670 9_testes_total |
V047 embryo |
SRR060689 160x9_testes_total |
SRR060663 160_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060657 140_testes_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060656 9x160_ovaries_total |
SRR060677 Argx9_ovaries_total |
V116 male body |
SRR060673 9_ovaries_total |
SRR060664 9_males_carcasses_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................GACCATCACGTCAATAGGC........................................................................................... | 19 | 3 | 7 | 1.86 | 13 | 0 | 0 | 8 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATG........................................................................................................................................... | 20 | 0 | 20 | 1.40 | 28 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGATGATGATGATGATGAGCGGA..................................................................................................................................... | 23 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GATGATGATGAGGGGAATGC................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................AATGACAATGATGATGATGCTGCT................................................................................................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................ATGATGATGAGGGGAATGCG................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATGA.......................................................................................................................................... | 21 | 0 | 20 | 0.95 | 19 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TGATGATGATGATGATGATGA............................................................................................................................................. | 21 | 0 | 20 | 0.65 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATGAGCGGA..................................................................................................................................... | 26 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TGACAATGATGATGATGTT................................................................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................AGACCATCACGTCAATAGAC........................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................ATGATGATGATGATGATGAT............................................................................................................................................ | 20 | 0 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GACCATCACGTCAATAGTC........................................................................................... | 19 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GATGATGATGATGATGATGAT............................................................................................................................................ | 21 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................ATTGCCAATGAAACTGAAGATGAT...................................................................................................................................................................................... | 24 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GATGAAGATGATGATGATGAGGG....................................................................................................................................... | 23 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATGAGGGGTTT................................................................................................................................... | 28 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GTTGCTGATGATGATGATGATGAT............................................................................................................................................... | 24 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GATGATGATGATGATGATGA............................................................................................................................................. | 20 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................TGATGATGATGATGATGATGAT............................................................................................................................................ | 22 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGATGATGATGATGAGGAGGGG...................................................................................................................................... | 22 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGATGATGATGATGAAGAGGG....................................................................................................................................... | 21 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................GATGATGATGTTGCTGATG............................................................................................................................................................................ | 19 | 0 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................GATGATGATGTTGCTGATG............................................................................................................................................................. | 19 | 0 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TGATGATGTTGCTGATGAT........................................................................................................................................................... | 19 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................CTGATGATGATGATGATGATGA............................................................................................................................................. | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................TGATGATGTTGCTGATGAT.......................................................................................................................................................................... | 19 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TGATGATGATGATGATGATGATGA.......................................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GATGATGATGATGATGAG......................................................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................ATGATGATGATGATGATGA............................................................................................................................................. | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GCTGATGATGATGATGATGA................................................................................................................................................ | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATGAGCGCA..................................................................................................................................... | 26 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................CGCTAAGGACAAGGTGTGA................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................AATGACAATGAAGATCATGGT................................................................................................................................................................................... | 21 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GCTGATGATGATGATGATGATGAT............................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGATGATGATGATGATGAG......................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GATGAAGTTGCTGAGGAT........................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGCTGATGATGATGATGATGAT............................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................ATGATGATGATGATGATGAG......................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................TGATGATGATGATGATGATGAG......................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................GATGAAGTTGCTGAGGAT.......................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GCTGATGATGATGATGATG................................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GATGATGATGATGATGATGAG......................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................CTGATGATGATGATGATGATGAT............................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGCTGATGATGATGATGATGATGA............................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................ATGATGATGATGATGATGATG........................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GATGATGATGATGATGAT............................................................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GCTGATGATGATGATGATGATGA............................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGCTGATGATGATGATGATGATGATG........................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TAACGGACGAGATACGACGGGCGTATTAACGGTTACTGTTACTACTACTACAACGACTACTACTACAACGACTACTACTACTACTACTACTACTCCCCTTACGCATACACTCACGACCGGTACTGGTACTGCAGTTATGTGTTTATGTTTGTGTAAATGCGTTATGCGTTCGGCCGTTGTTCGTGCTGTTGTAACTGCGATTACTGCTCCACAGTTGCCGTGCACTTGCTGT
**************************************************((((((.............((.((.((.((....)).)).)).)).((..((((((((((((((.((....((((......))))...............)))))))))...)))))))..))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060668 160x9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060656 9x160_ovaries_total |
SRR060666 160_males_carcasses_total |
M027 male body |
SRR060667 160_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060679 140x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060654 160x9_ovaries_total |
SRR060670 9_testes_total |
SRR060688 160_ovaries_total |
V053 head |
SRR060662 9x160_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................TTCGTCAGGTGTTCGTGCTG............................................ | 20 | 3 | 9 | 0.89 | 8 | 1 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGTCCGGTGTTCGTGC.............................................. | 19 | 2 | 4 | 0.75 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGTCCGGTGTTCGTG............................................... | 18 | 2 | 14 | 0.71 | 10 | 3 | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGTCTGTTGTTCGTGCTG............................................ | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGTCCGGTGTTCGTGCGG............................................ | 21 | 3 | 6 | 0.50 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGGCAGGTGTTCGTG............................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AACGACTACTACTACTACTA.................................................................................................................................................. | 20 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TTCGGCAGGTGTTCGTGC.............................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AAGGACTACTACTACTACTACTACT............................................................................................................................................. | 25 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GGACGAGTTACGACCGGCG................................................................................................................................................................................................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CAACGACTACTACTACTAC.................................................................................................................................................... | 19 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................GTTCGGCGGTCGTTCGTG............................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................ACGTTCGGCGGTCGTTCGTG............................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CGTTCGTCAGGTGTTCGTGC.............................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CGTTAGGCGGTTGTTCGTGG.............................................. | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................GACTACTACTACTACTACTACT............................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................TACTACTACTACTACTACT.......................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CTACTACTACTACTACTACTACT.......................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................ACTACTACTACTACTACTACT.......................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ACTACTACTACTACTACTACTACT.......................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................ACTACTACTACTACTACTCGC....................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................CGACTACTACTACTACTACTACT............................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_10324:935013-935244 + | dvi_826 | ATTGCCTGC---------------------------------TCTATGCTGCCCGCATAATTGCCA---------------------------------------------------------ATGACAAT--G----------------------------------------------------------ATGATGATGT------TGCTGATGATGATGTTGCTGATG---ATGAT---------GATGATGATGATGAGGGGAATGCGTATGTGAG--------------------------------------------TGCTGGCC------------------ATG-------------------------------------------------ACCATGA------C-----------------------------------------------GTCAA---TACACAAATACAAACAC------------------------------------------ATTTACGCAATACGCAAGCCGGCAACAAGC--------------------------------------------------------------------------------------------------------------A------------CGACAACATTG--------------------------------------------------------ACGCT--------------------------AAT------GACGAG-------------------------G-TGTC-----------AACGG---------CACGTGAACGACA |
| droMoj3 | scaffold_6496:2807833-2808022 + | ATTGCCTGC---------------------------------GCTATGCTCGACGCATAATTGTTG---------------------------------------------------------ATCATGAT------------------------------------------------------------GATGATCGTGT------TGGTCGTGATGATGTTGCT--------------------------------------------------------------------------------------------------GCTGGCC------------------ATG-------------------------------------------------ACCGTGA------C-----------------------------------------------GTCAA---TACACATATACTAGCAC------------------------------------------ATGCACGCAATACGCACGCCGGCAACAACA--------------------------------------------------------------------------------------------------------------A------------CGACATCATTG--------------------------------------------------------ACGCT--------------------------AAT------GATGAG-------------------------G-TGTC-----------AACGG---------CACGTGAACAACA | |
| droGri2 | scaffold_15245:8616628-8616892 + | ATTGCCTGC---------------------------------GTTATGCTGGCCGCATAATTGCCA---------------------------------------------------------AAGCCAAT--G----------------------------------------------------------ATGATGATGA------TG---------------------------------------ATGATGACGATGATGGGAATGCGTATGTGAA--------------------------------------------TGCTGTCC------------------ATG-------------------------------------------------ACCATGA------CGTCAATGTGG--CCGCAGTGA--------------------GACAATC----------CCAAATACAACCACACTCACACTCAGATTCACACTCGCACTCGCACTCACATTCATATTCACGCACTACGCACGCCGGCAACGACA--------------------------------------------------------------------------------------------------------------A------------CGACAACATTG--------------------------------------------------------ACGCT--------------------------AAT------GACGAG-------------------------G-TGTC-----------ATCGG---------CACGTGAACAACA | |
| droWil2 | scf2_1100000004830:291923-292161 - | TGTGCCTG-ATAT---------------------------------TGCTGCTGCTGCTGCTGTTG---------------------------------------------------------CTCCCAAT------------------------------------------------------------GACAATTATGAGAATGATA------ATAATGATG---------ATG---------ATGACGATG------------GG------------------------------------------------GACAACA--------------------------------ACC--------AAA--------------------------------AGA---AC-----------------------------------------------------------------------------------------------------------------------------------------AATAT--------GGAT------GACAA----CAACTCGTCGTCGGAACAACAGCCCGATATTATACCATATCCACTTAGTGTGGGTGAAACAAGCTGGAAACCCGCAA-AACTACAGC---AACAGCAATGGCAACAGCAACA---------------------------------------------------TCA------------------------------------------------------------------------------------T----------------------CACGAAAATGACA | |
| dp5 | XR_group6:3859602-3859793 - | ATTGCTGCT--G------------------------------------CTGCTGGTGTTGTTGTTG---------------------------------------------------------TTGTTGCT------------------------------------------------------------GGTGGTGGTGT------TGCTGATGTTGTTGCTGCTGCTG---CTGCTGCTGATG---------ATGATGA------------------------------------TGGT-------------G--------------------------------------------------------------------------------------------------GTGCTGCTGATG--CTGCGGCGAA-------------------GACTGTCCGGA---CATGGGTAT---------------------------------------------------------------------------AGGCA---------------------------------------------------------------------------------------------------------------------------------------------------------GCGGCGAGCTGGGG-----CATCC-----------------T--------------------------AAT------GGCGAG-------------------------GGTATTGGCGAGGCGACAAT----------------------GG | |
| droPer2 | scaffold_9:2150861-2151055 - | ATTGCTGCT--G-------------------------------C--TGCTGCTGGTGTTGTTGTTG---------------------------------------------------------TTGCTGCT------------------------------------------------------------GGTGGTGGTGT------TGCTGATGTTGTTGCTGCTGCTG---CTGCTGCTGATG---------ATGATGA------------------------------------TGGT-------------G--------------------------------------------------------------------------------------------------GTGCTGCTGATG--CTGCGGCGAA-------------------GACTGTCCCGA---CATGGGTAT---------------------------------------------------------------------------AGGCA---------------------------------------------------------------------------------------------------------------------------------------------------------GCGGCGAGCTGGGG-----CATCC-----------------T--------------------------AAT------GGCGAG-------------------------GGTATTGGCGAGGCGACAAT----------------------GG | |
| droAna3 | scaffold_12088:3486-3685 - | TGACGATG-ACGA---------------------------------------------------------------------------------------------------------------------T--GA----------------CGATGACGATGACGATGACGATGACGATGACGATGACGATGACGATGACG------------------------------------------------ATGAC---------------------GATGA--------------------------------------------CGATGACGATGA------CGATGGCGATGACG-----------ATG-----------------------ACG------ATGACGA------TGACGATG--------------------------------------------------ACGATG-A-----------------------------------------------------------------CGATGA--------CGAT------GACGA------------------------------------------------------------------------------------------T------------------------------------GACG-------ATGACGATGACGAT-----GACGA-----------------T--------------------------GAC------GATGAC-------------------------GATGACGATGA----------------------CGATGACGATG | |
| droBip1 | scf7180000396022:3474-3711 - | TCGCAGCA-GCAG-----------------------------------CAGCA-------------------------------------------------------------------------------------------GCA-------------------------TGAAGACGATGATGAAGACGATTACGATGA------TG------------------------ATG---------AAGAGGATG------------AGGAA---GATGAGGATCATCATCAGGATCAGG-------------GTAATGGACAC--------------------------------GAT--------CTA-----------------------ACG------------C-------------------------CCAC-------------AGATCTGCGGCACATGA-----TACACG-A------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCGCAGCAGCAGCAAC---AGTCGCAG-----------------------------------------------------------------------------CAGCAGCAGCAGCAACAGGATCCCATCGACTACGAGGACGAG--GACGAC-----------------GCCGACGACGA----------------------CGAGGACGATG | |
| droKik1 | scf7180000302472:3238233-3238440 + | TGTGGATG-CTG-----------------------------------------------------------------------------------------------------------------------ATGT------TACTG----------------------------------------------ATGCCTCTGC------TGCTGC---------------TA---CTGCTGCTGACGATGATGATG---------------------------------------------ATGATGATGCGAC-----------TGCTGAAA------------------------GCT--------CCC-----------------------ATG------------A-------------------------CATTGAGCTGCG-----------------GT-GCCAAGCGACGACT-------------------------------------------------------------------------------------------ACAATGAGCAATTT----------------------------------------------------------------TTCGTA--GCTTCCAAGCTTGTTAACAGGCGCAATGACAACG---------------------------------------------------TCAGACAGATGAT--------------------------GAT------GACGAC-------------------------G-------TTGCA-----------------------CGTCAACG | |
| droFic1 | scf7180000453549:133520-133745 + | AGTGCGTCC---------------------------------ATTATGTGAGCGA---GCTTAATTTGAATACGATTTTTGGTCTTCTCTACTTGCAGCTAAAGGCCA-------------------------------------------------------------------------------------------------------------------ATCGAATG---CTGTGGGAGAAT---------AAGGCCA------------------------------------AGCA-------------A--------------------------------------------------------------------------------------------------GTGCTGCAGATGTCCAGCGAGGAT-------------------GACAACCTCAA---CACGATTAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAC---A-------------------------------------------ATAGCAA----------------------------------CAATAAGGATAACAAC-----------AAT---AAT--CAGTACACAATCAATAAAATTAT-------ACCAGAACAA----------------------CGCGGCAGACA | |
| droEle1 | scf7180000490214:547459-547688 - | ACAACAACA---------------------------------GCAATACTAGCAGCAACATTAGCA---------------------------------------------------------GCAACTCT------------------------------------------------------------GGCGACAACAC------AGC----------------------------------------------------------------------------AACGGAAGTGGCC-------------ACGAAATGTGA--------------------------------GTACGCGCAACTCGAGCGTGAGAGGCAGAGCGTGAGCGCAGGCGCA-TA---C--------CTG----CAACCACCCAATGT-------------------GAC-----AAT------CAGCA-G-----------------------------------------------------------------CACCAGCAGCAGC--------------------------------------------------------------------------------------------------AGCAACAAC---AA------------CAACAA-----------------------------------------------------------------C--------------------------AAT------AACGGG-------------------------AGCAAC-----------AACAAGGGGAGCAGCGCCTGCTCAATA | |
| droRho1 | scf7180000766551:36405-36589 + | TACAAATG-ATG-----------------------------------------------------------------------------------------------------------------------ACGA------TGAAGA-------------------------TGTAGATGAAGATGAAGATGACGATAATG------------------------------------------------CAGATG------------AAGATAAT---------------------------GAGGATG--------------------------C------AACTGGGGAAGAAA-----------ATG-----------------------GTG------ACGAAGA------CGAGAACG--------------------------------------------------AAGAAG-A-----------------------------------------------------------------CGAAAA--------CGAC------GATGA------------------------------------------------------------------------------------------C------------------------------------GAGA-------ACGACGATGACGAG-----AACGA-----------------C--------------------------GAT------GACGAG-------------------------AACGACGATGA----------------------CGAGAACGACG | |
| droBia1 | scf7180000302113:3803943-3804184 + | GGAAGTCG-ATT---------------------CCA------------CGAGGCGCGTATTAGTCG---------------------------------------------------------ATG-----ATGC------T-------------CATTATGAGGATGCC------------------------------GC---------------------TGCCGCTGACGACG---------ATGACGATG------------AC---------------------------------GTTG-------CGCGGCCGCCG--------------------------------GTT--------ATA--------------------------------GAA---ATAGAAA---------------------------AGCG-----------------GCAGC---CAAAT----A-------------------------------------------------------------------------------CATCAACAACGAAAACGGAAAAA------------------------------------------------------------------TCAACAGTGGCAATAGCAGCAGCAGCAGCAGCAACAGCAACA---------------------------------------------------TCGT-----CTT-----------------------------T---GGG--AGACGCGTCATCGCCAGTGTTGTCCCTGT-----------------------------------CATCCGAAA | |
| droTak1 | scf7180000415381:755652-755894 + | ATGATATG-GAG------------------------------------CTGGATGCACAGCAAGAA--------------------------------------ACCAGAACTGAGCCAAGTGCAA-----CAAC------G-------------CAACAGCAGGATCTT------------------------------GC---------------------CGATCTTGAGGAGC------------AGAACAAGGATGCGGGGGAGGCTAGTTTAAA--------------------------------------------TGTTAGCAATAATCACACAACCGATAGCAATA----------------------------------------------ATAGTTG------TAGTCGAA--------------------------------------------------A------------------------------------------------------------------------------------G--------------------------------------------------------------------------------------------------------------AA------------CAACAATGGTGGCGGCAATGAGAGTGAGCAACATGTT------------GCCAA-----------------T--------------------------TCG------GCGGAG-------------------------GACGATGATTG----------------------CGCCAACAACA | |
| droEug1 | scf7180000409711:2042976-2043205 - | ACCGGCAAC-ACTTGTTGCTGCCGGTGTTGTTGCTCCGGGGAATGTTGCTGCTGGCACCGTTGACA---------------------------------------------------------CGGACA--ACGCTGACGATGACG-AATATGATGAGGACGAC------------------------------------GA---------------GGACGATGATGATGATGATA---------TTGATGATG------------GA------------------------------------------------G-------------------------------------------------TCG-----------------------ATG------AAATTGG------T-----------------------------------------------GAAAT---TGCAGGTA-T-----------------------------------------------------------------CGCCGGTGAC--------------GACGACGAGGAGG------------------------------------------------------------------CCGACGATGA-----------------------------------------------------------------------------------------------T--------------------------GAT------GACGAT-------------------------GACGACGATGA----------------------CATTATCGGCG | |
| dm3 | chr3R:13608769-13608995 - | GGAAGTCG-ATT---------------------CGA------------CGAGGCGCGTATTAGTCG---------------------------------------------------------ATGCTCAT--T----------------------------------------------------------ATGAGGATGC------CG------------------------CTGAC---------------------------------------------GACGACGATGACGTTG-------------TGCGGCCGCCG--------------------------------GTT--------ATA--------------------------------GAA---ATAGAAA---------------------------AGCG-----------------GCAGC---CAAAT----A-------------------------------------------------------------------------------CATCAACAACGAAAACGGAAAAA------------------------------------------------------------------TCAACAGAGGCAGCAACAGTAGCAGCAGCAGCAGCAGCAACA---------------------------------------------------TCAG-------------------------------------T---GGG--AGACGCGTCATCGCCAGTGTTGTCCCTGT-----------------------------------CATCTGAAA | |
| droSim2 | x:12068153-12068329 + | ATGAA-TG-ATG-----------------------------------------------------------------------------------------------------------------------ACGA------TAACG----------------------------------------------ATGACGATGA------CGATGATAATGATGATGATGATG---ACAAT---------------------------------------------TACAACCGTGATGATG-------------GCGAATCATCG--------------------------------GAT--------TGA-----------------------ACG------------A-------------------------CATT-------------------AGCTTGGCCAA---CAAACAAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAACAAATTGACCAAGCCGGCAGCACCAACATCAG----------------------------------------------------------------------------ATATTAATTACACGTCAAC----------------------GG | |
| droSec2 | scaffold_21:464171-464347 + | ATGAA-TG-ATG-----------------------------------------------------------------------------------------------------------------------ACGA------TAACG----------------------------------------------ATGACGATGT------CGATGATAATGATGATGATGATG---ACAAT---------------------------------------------TACAACCGTGATGATG-------------GCGAATCATCG--------------------------------GAT--------TGA-----------------------ACG------------A-------------------------CATT-------------------AGCTTGGCCAA---CAAACAAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAACAAATTGACCAAGCCGGCAGCACCAACATCAG----------------------------------------------------------------------------ATATTAATTACACGTCAAC----------------------GG | |
| droYak3 | X:20462015-20462197 - | TGGCGATG-GCC---------------------------------------------------------------------------------------------------------------------AT--GA----------------TGATGACGATGAT------------------------------------GA---------------TGACGATGATGATGACGATG---------ATGACGACG------------AT---------------------------------GATGATG-----TGATTCATGT--------------------------------GAT--------TCA----------------------------------T---GCGGAAA---------------------------ACAAGCAACAATTTTGGTTCACCAGA-----CAGACA-A-----------------------------------------------------------------CGCCGGC--------------------------------------------------------------------------------------------------------ACCAACAAC---AAAGGCAG-----------------------------------------------------------------------------C--------------------------AAT------AACAAG-------------------------AATATC-----------AGAAG---------GGGATAAACAACA | |
| droEre2 | scaffold_4690:17812978-17813168 - | CACTGGCA-ACGA---------------------AA-----------ACTGG------------------------------------------------------------------------CC-----ATGA------TG-------------------------------------------------ACGATGACGA------TGGCGA---------TGATGATGATGAGG---------ATGCGGATG------------AGGATGAT---------------GAGGATGCAA-------------GTGAT-------------------------------------------------TCA----------------------------------T---GCGGAAA---------------------------ACAAGCAACAATTTTGGTTTACCAGA-----CAGACA-A-----------------------------------------------------------------CGCCGGC--------------------------------------------------------------------------------------------------------ACCAACAAC---AAAGGCAA-----------------------------------------------------------------------------C--------------------------AAT------AACAAG-------------------------AATATC-----------AGAAG---------GGGATAAACAACA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:16 PM