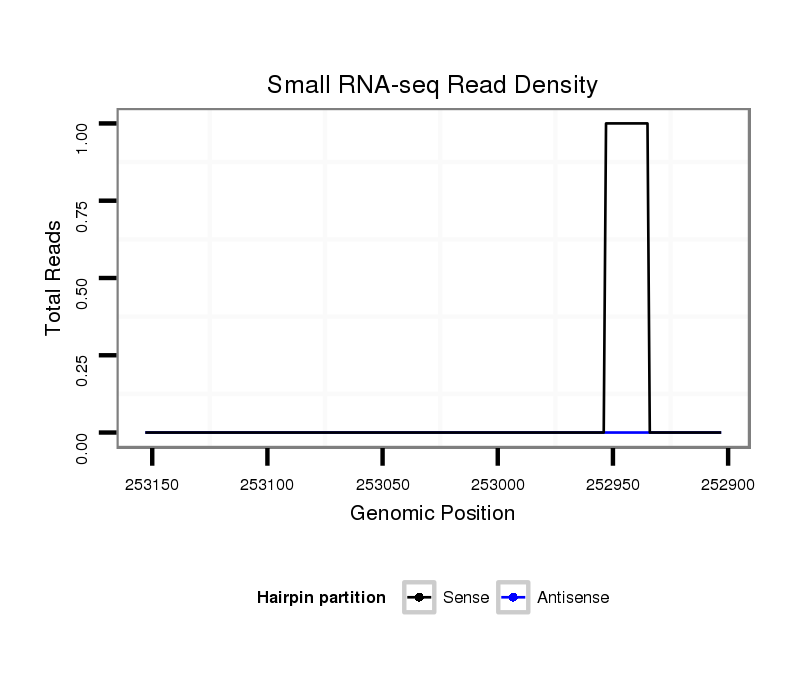
ID:dvi_8212 |
Coordinate:scaffold_12928:252953-253103 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ16692-cds]; exon [dvir_GLEANR_17195:2]; intron [Dvir\GJ16692-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAAGTCTGGCTAGATAAATTGCCTAATTAATTCAGTTTCTTTGGCCGCTGAAAAAGTAGAGTCCTGGCTCGTAACAATACGATTCTAGCCTTAGCTTAAAATAACATATATCATAAATATTTTCTACTCTTAAGGTATTACAAAATCTTTCCGAAGAGAAGGACATTACATTAAAATTACTATCAATCCCAACTCTTCCAGTTCCTGAGTGCGCTGGGCCAAGTGAACAGCCCAGGAGCAGCTGTTAACCC **************************************************.....((((.(((((((((((((....))))).....)))....................................((((((((((........)))))....))))).))))).))))................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
SRR060656 9x160_ovaries_total |
V053 head |
V116 male body |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................GTTCCTGAGTGCGCTGGGC................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TGAGCCGCTCAGGAGCAGCTG....... | 21 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................TAACATAAATATTCTCTATTCT......................................................................................................................... | 22 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................GAAGGAGATTACGTCAAAAT.......................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................TATTGAAGAATCTTTCCGA................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TTTCAGACCGATCTATTTAACGGATTAATTAAGTCAAAGAAACCGGCGACTTTTTCATCTCAGGACCGAGCATTGTTATGCTAAGATCGGAATCGAATTTTATTGTATATAGTATTTATAAAAGATGAGAATTCCATAATGTTTTAGAAAGGCTTCTCTTCCTGTAATGTAATTTTAATGATAGTTAGGGTTGAGAAGGTCAAGGACTCACGCGACCCGGTTCACTTGTCGGGTCCTCGTCGACAATTGGG
**************************************************.....((((.(((((((((((((....))))).....)))....................................((((((((((........)))))....))))).))))).))))................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
V116 male body |
SRR060687 9_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
SRR060689 160x9_testes_total |
V047 embryo |
SRR060660 Argentina_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................AGCGTTGAGAAGGTGAAGG.............................................. | 19 | 2 | 6 | 8.33 | 50 | 22 | 10 | 6 | 0 | 3 | 5 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AGCGTTGAGAAGGTGAAG............................................... | 18 | 2 | 10 | 1.20 | 12 | 4 | 3 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTTCATTTCAGGACCAAGC.................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................GCGTTGAGAAGGTGAAGG.............................................. | 18 | 2 | 15 | 0.73 | 11 | 2 | 4 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AGCGTTGAGAAGGTGAAGGTC............................................ | 21 | 3 | 7 | 0.57 | 4 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AGAAGGTTAAGGACTCAAACG..................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GTTGAGAAGGTGAAGGTC............................................ | 18 | 2 | 14 | 0.29 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 |
| ..........................................ACCGCGACTTTTTCATCT............................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TAATGATCGCTAGGGTGGA......................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................AAACCGCGACTTTTTCATCTA.............................................................................................................................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................GTCGCAGGACCTAGCATTG................................................................................................................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................AGGTTGAGAAGGTAAAGGA............................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................ATAGCGTTGAGAAGGTGAAG............................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TAATGATCGCTAAGGTTGA......................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:252903-253153 - | dvi_8212 | AAAGTCTGGCTAGATAAATTGCCTAATTAATTCAGTTTCTTTGGCCGCTGAAAAAGTAGAGTCCTGGCTCGTAACAATACGATTCTAGCCTTAGCTTAAAATAACATATATCATAAATATTTTCTACTCTTAAGGTATTACAAAATCTTTCCGAAGAGAAGGACATTACATTAAAATTACTATCAATCCCAACTCTTCCAGTTCCTGAGTGCGCTGGGCCAAGTGAACAGCCCAGGAGCAGCTGTTAACCC |
| droMoj3 | scaffold_6359:1595204-1595240 - | TG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTCAACCCACAAATCCTCTAGCAGCTGGAGCACA | |
| droGri2 | scaffold_15203:6716789-6716849 - | CAATTCCA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGTTCCTTGTTGCTCTGAGCCAAATGAGCAATGCTGGCTTCCTTGGAGCCCC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:43 PM