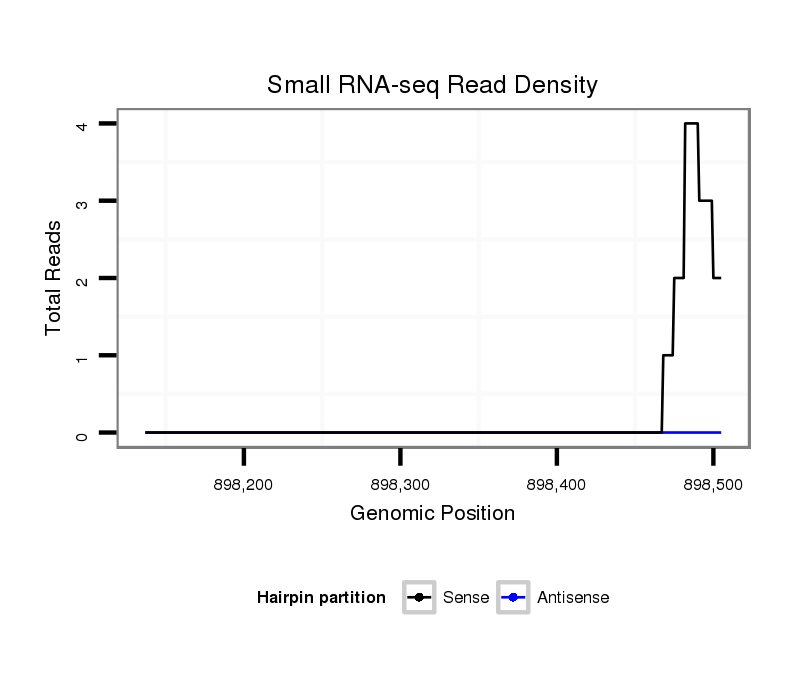
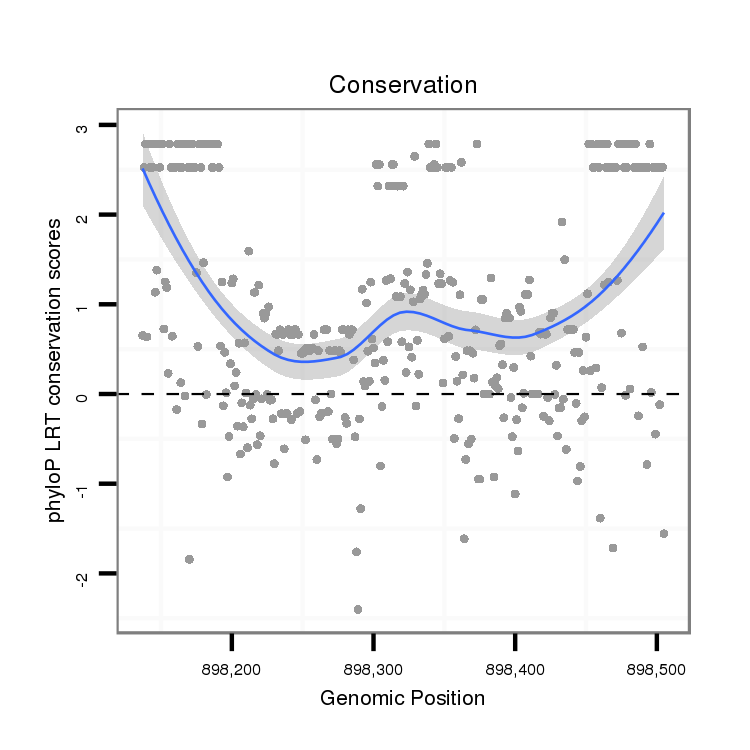
ID:dvi_821 |
Coordinate:scaffold_10324:898187-898455 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ15108-cds]; exon [dvir_GLEANR_15408:3]; CDS [Dvir\GJ15108-cds]; exon [dvir_GLEANR_15408:2]; intron [Dvir\GJ15108-in]
| Name | Class | Family | Strand |
| (CAAAC)n | Simple_repeat | Simple_repeat | + |
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CGTCTGCCATTTGATTGCCTCGCCGACCGTCTCGCTGCAGAGCAACAAAGGTAAGTTGGCGTTGCTCCCATCCCCATCCCCAACCACCACCTATCCAAGCGTATCTGATTGGCCGTACCATCGAATTAAAAGCGGAATCTGAATCTGAAACTCCAAATTGTAATCTGTGTATAGCTAACCGGGCGTATGCGTAATATTCATTTGCCTTGTGCTTGCCGCCAAACAAACCAAACCAAACCAAACAAAACGCCTGCGCCTGCCTTCATTCTTTATTCTTTTGTCTTCCGTCAATTAATTCTTATAATTCGAATATTCACAGCCTCCTTGGATCGGCCCAAGACGGAAATGTCGCGCGCCGAGGCCGCTGCG **************************************************....(((((...((..........))...)))))..........((((.((....)).))))........((((((((.(((((((.....(((.((......)).)))....))))............((((((((.((((........((((..(((((.(((....))).))...)))..)))).........)))).))))))))......................................))).))))))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
M047 female body |
GSM1528803 follicle cells |
SRR060681 Argx9_testes_total |
SRR060666 160_males_carcasses_total |
M028 head |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060660 Argentina_ovaries_total |
SRR1106728 larvae |
V047 embryo |
V116 male body |
SRR060656 9x160_ovaries_total |
SRR060680 9xArg_testes_total |
SRR1106727 larvae |
SRR060657 140_testes_total |
SRR060665 9_females_carcasses_total |
SRR060683 160_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................AGCGGACTCTGAATC................................................................................................................................................................................................................................ | 15 | 1 | 9 | 2.56 | 23 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................ATGTCGCGCGCCGAGGCCGCTGCG | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................GGCCCAAGACGGAAATGTCGCGC............... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................GACGGAAATGTCGCGCGCCGAGGCC...... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACAGCCTCCTTGGCTGGG.................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACAGCCTCCTTGGGTGGG.................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................AAAGCCTCCTTGGATCGGCCCAAGAC............................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................AAAGCCTCCTTGGATCGGCCCA................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................CGGAAATGTCGCGCGCCGAT......... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................CAAAGCCTCCTTGGATCGGC................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CTTTGGATAGCTTACCGGGC......................................................................................................................................................................................... | 20 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CATGGAATTAATAGCGGAATCC..................................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................CTTGGACCGGCGCAAGACGGA......................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................CTTGGACCGGCGCAAGACG........................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACAGCCTCCTTGACTGGG.................................... | 20 | 3 | 8 | 0.38 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CATTTAATTCCCTCGCCCACC..................................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................ACGAGGATCGGCCCAAGACGG.......................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CAGAGCAACAAAGGCAAATT........................................................................................................................................................................................................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................CTTGGACCGGCGCAAGACGGAC........................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................TGGATCGTCGCAAGACGGAC........................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AAAGCGGAATCTGACTGT............................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................TGGACCGGCGCAAGACGGA......................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACAAGGGGAATCTGACTCT............................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.25 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................CTTGGATCGTCGCAAGAC............................ | 18 | 2 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACATCCTCCTTGGTTTGGC................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCGCAGCCTCCTTGGGTGGG.................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................GGATCGGCGCAAGACGGACC....................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TCTGATTGGACTTACCAT........................................................................................................................................................................................................................................................ | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACAGCCTGCTTTGATCTG.................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TTTCTTCTTTTGTCCTACGTC................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................TCTTGGACCGGCGCAAGACG........................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TCGCCGAACGTCTGGCGGC........................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TTTGTTCTTTTGTCTTACGTTA............................................................................... | 22 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................TCACAGCGTCCTTAGATCT..................................... | 19 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TTACCGGCCGTATGCGGAA............................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AGAGCAACAAAGGGAAGTCGG...................................................................................................................................................................................................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................AGAGCAAGAAAGTTATGTTGG...................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................CTACAAAGGTAACTTGGCT.................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................AAGGGGAATCTGACTCTC.............................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCAGACGGTAAACTAACGGAGCGGCTGGCAGAGCGACGTCTCGTTGTTTCCATTCAACCGCAACGAGGGTAGGGGTAGGGGTTGGTGGTGGATAGGTTCGCATAGACTAACCGGCATGGTAGCTTAATTTTCGCCTTAGACTTAGACTTTGAGGTTTAACATTAGACACATATCGATTGGCCCGCATACGCATTATAAGTAAACGGAACACGAACGGCGGTTTGTTTGGTTTGGTTTGGTTTGTTTTGCGGACGCGGACGGAAGTAAGAAATAAGAAAACAGAAGGCAGTTAATTAAGAATATTAAGCTTATAAGTGTCGGAGGAACCTAGCCGGGTTCTGCCTTTACAGCGCGCGGCTCCGGCGACGC
**************************************************....(((((...((..........))...)))))..........((((.((....)).))))........((((((((.(((((((.....(((.((......)).)))....))))............((((((((.((((........((((..(((((.(((....))).))...)))..)))).........)))).))))))))......................................))).))))))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V116 male body |
SRR060658 140_ovaries_total |
V047 embryo |
SRR1106728 larvae |
SRR060676 9xArg_ovaries_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
GSM1528803 follicle cells |
SRR060667 160_females_carcasses_total |
V053 head |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................CAACCGCTTCGAGGGTAG......................................................................................................................................................................................................................................................................................................... | 18 | 2 | 3 | 1.00 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................TTACAGACGCGGAAGGAAGTA....................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGATTATCGCCTTAGAGTTA................................................................................................................................................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CGGAACACGAAAGGCGGTC................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CACACATCGATGGGCCCG......................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................AGAAGGCAGTGAAGTGAGAA..................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CGATGGTAGTGGTAGAGGTT.............................................................................................................................................................................................................................................................................................. | 20 | 3 | 16 | 0.13 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGGTTGGTTTGGTTTGGTTTG.............................................................................................................................. | 21 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TCTACCTCAACGGGGGTAG......................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TTTGGTTTGGTTTTGTATGTT............................................................................................................................ | 21 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GGAGCGGCTGGCCGA................................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................TTACAGACGCGGAAGGAAGT........................................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................AGGAAGTTAATTAGGACTAT.................................................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................GGAAGTTAATTAGGACTATT................................................................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AACAAGAGTAAGGGTAGGGGT............................................................................................................................................................................................................................................................................................... | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................GCATCGTAGCTTAATTTGG............................................................................................................................................................................................................................................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GCGCATCGTAGCTTAATTT............................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TAACCGGGATGTTAGCTTC................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_10324:898137-898505 + | dvi_821 | CGTCTGCCATTTGATTGCCTCGCCGACCGTCTCGCTGCAGAGCAACAAAGGTAAGTTGGCGTT-----------G---CTC-----------------C-----CA-------------TC--CC--C-A----------TCCCCAA---------CCACCAC-------CTA-------TC------------------------------------------------CAAG----CGTATCTGATTGGCCGTACCATCGAATTAAAAGCGGAATCTGAATCTGAA---------------------------------------------A-------CTCCAAAT----TGTAATCTG----------TGTA--TAGCTAACCGGGCGTATGCGTAATATTCAT----TTGCCTTGTG--CTTGCCG-C---------------------------CAAA-------CAAACCAAACC-----A----AA-----CC-AAACAAAA-CGCC--TGCGCCTGCC------------------------T------TCATTCTTTAT---------------------------------------------------------------T-CTTTTGT-CTTCCGTC-AA---------------------------------------------------------------------------TTAATTCT------TATAAT--TCGA---AT-ATTCACAGCCTC-CTTGGATCGGCCCAAGACGGAAATGTCGCGCGCCGAGGCCGCTGCG |
| droMoj3 | scaffold_6496:2769076-2769523 + | CGTTTGCCATTTGATTGCCTCACCGACCGTCTCGCTGCAGAGCAACAAAGGTAAGTTGGCATT-----------G---CTC-----------------------CG-------------AT--CTCCG-A----------TCTCCGA---------TCACCA---AAC----T-------C--------------------------CGAC-TTCCGATCTCCATC----CAAAATCCCCTATCCGATTGGCC-------CACACTAAAAG--AAAATTGAAAATGAAAAACAAAAGCACCTCGAACCTA-----------------------------------CAAAATTGTGTAATATG---------TTTAC--TTGCTAACCGGGCGTATGCGTAATATTCAT----TTGCCTTGTG--CTTGCCAGC---------------------------CAAAAACCCCACAAAAAATATCAAAATAAATAAAAAGAAAAG-------AAATCCCAC--G------------------------CA--ATA---------------------------CCTAAGCCTGCGACTGCGCCTGCGCCTGCACCTGCCTCAGCTTCCAGTTGGAAT-----------------------------------------------CTTTTCT------------CATTGAATC------------------------------------------TTTGT-ATG-TTTCACAGCTTC-CTTGGATCGGCCCAAGACGGAGATGTCACGCGCAGAGGCAGCTGCG | |
| droGri2 | scaffold_15245:8585976-8586377 + | CGTCTGCCATTTGATTGGATCACCGACCGTCTCCCTGCAGAGCAATAAAGGTAAGTTGGTGTT-----------G---CTCCAAC--CTTTTG--GCAACCCTCCA-------------CT--CC------------------------TGCAAGTGC--CC-CTGCC----ATGTAACAC--------------------------CTATCCCTTCCCTACCACCAATCC-AG----CGATTCCGATAGA--GAACCATCA--TAGAAAAC--------AATCTGAA---------------------------------------------A-------TT-------ATAAATAATCTG-----TA---TATATAGAGCTAACTGGGCGTATGCGTAGTAT-----TCATTGCCTTGTGCTTGTGCCG-C---------------------------CAAA---------AAT----------TG----AATAAAACCAAA-----A-CGCC--TGCGCCTGCC------------------------T------TAGGTCCT-----TTGCTGAC-------------------------------TTCCCA-------------------------------GCCTAACTAATCGA---GACCTTCCCCGCCGA-----------------------------------------------------------------------------ACT-TTTCACAGCATC-CCTGGATCGTCCCAAGACGGAAATGTCGCGCGCCGAGGCCGCTGCC | |
| droWil2 | scf2_1100000004510:1314703-1315093 - | CGTTTGCCATATGATTGCCTCGCCAACGGTCTCACTGCAAAGTAATAAAGGTAAGCTGGGCGT-----------G---CATCAGAAAAACGAA-----------AGACGATACGACATAGCGACC----AATTCCACAAAATC------CAACTAGG---CC-CAGTTAATCC-------T-------------CCAGATGCAGTC---CT---------------------------------------------------------------------------------------------------TTCCAGCTTCCATATAAGC---------CTT----------------------------------------------------CTGCCTACTGCCTGCCTGCCTGCC-TGCT--TGTGCTG-C---------------------------CCTT-------ACAGA-----------A----AAAAA------------TATCGC--TGCGGCTGCTTATTCTATTTACTTTCTATATGATATATAATATA-----TATATTTCTTGA--------------------------------------------------------------TCCTCCCATC-AA----T----------AGATGATCTAAGATTTACT--TAAAATA-----------------------------TG-----------------------TCGAT-TTT-TCTCATAGCTTC-TTTGGATCGTCCCAAAACGGAAATGTCACGTGCAGAAGCTGCCGCA | |
| dp5 | 3:543833-544189 - | dps_914 | CGTCTGCCACATGATTGCCTCGCCAACGGTCTCCCTGCAGAGTAACAAAGGTAAGCTTCGTGCAGAG--CCTGGGGCG---------CACAAG--GTCA-----CA-------------AT--TT--G-A----------TTTCCCA---------TTACCAC-------TTT-------TCCGTAACGTAGCCGTAACCAGAGTCC------------------------------------------------------------------------------------------------------AAGAGAA-----------ACCAATA-------TGTATATCCGTGAGAAATGTG----------TGAC--AAACTAACCGGGTGATTGCTCACTA-CCGTTTCATTGCCTTGCT--TTAGCCG-C---------------------------CTTC-------AAAGC----------AG----CA-------------------------------------------------------------------------------------------------------------------CCTGTCA----------------------------------------GTCGAGCAGGTCTGCCGTTCCAAACTTTTACTATAAAATG-------------------------------T--------------A------TCAT-TTTCT-TCTGATAGCTTC-ATTGGATCGCCCCAAGACGGAAATGTCGCGCGCAGAAGCCGCGGCA |
| droPer2 | scaffold_2:708853-709210 - | CGTCTGCCACATGATTGCCTCGCCAACGGTCTCCCTGCAGAGTAACAAAGGTAAGCTTCGTGCAGAG--CCTGGGGCG---------CACAAG--GTCA-----CA-------------AT--TT--G-A----------TTTCCCA---------TTACCAC-------TTT-------TCCGTAACGTAGCCGTAACCAGAGTCC------------------------------------------------------------------------------------------------------AAGAGAA-----------ACCAATA-------TGTATATCCGTGAGAAATGTG----------TGAC--AAACTAACCGGGTGATTGCTCACTA-CCGTTTCATTGCCTTGCT--TTAGCCG-C---------------------------CTTC-------AAAGC----------AG----CA-------------------------------------------------------------------------------------------------------------------CCTGTCA----------------------------------------GTCGAGCAGGTCTGCCGTTCCAAACTTTTACTATAAAATG-------------------------------T--------------A------TCAT-TTTCT-TCTGATAGCTTCATTTGGATCGCCCCAAGACGGAAATGTCGCGCGCAGAAGCCGCGGCA | |
| droAna3 | scaffold_13266:7570201-7570538 - | dan_1868 | TGTCTGCCACATGATAGCATCGCCCACCGTCTCCCTGCAGAGCAACAAAGGTAAGGCCGCACC-----------GGAG---------TACAGGAGTTTT-----CT-------------CA--TC------------------------TGA----------------------------------------CCCGAACCCGAGGCC------------------------------------------------------------------------------------------------------CTGCGAGCATCTACG--------------------TGATCCGC-ATCAGAGTG----------TCAA--TAACTAACCGGGTGACTTTGCACTACCCGTATCATTGCCTTGCT--TCCGCTG-C---------------------------CTTC-------CAAAA----------AG----AAAGG----GAA-----ATCCTT--CAT----------------------------------------------------------------------------------------------------TTCCCGCTGGAATCC-------------------------------------------------CTT------------CA-----TTCGTTCAAC-CCTTTTCATGCTTGCCTTTC-----T------CTTAAT-TGCCCCCGACAGCTTC-TCTCGACCGCCCCAAGACGGAAATGTCACGGGCCGAGGCGGCCGCA |
| droBip1 | scf7180000396759:1246379-1246724 + | TGTCTGCCACATGATAGCATCGCCCACCGTCTCCCTGCAGAGCAACAAAGGTAAGGCCGCCCC-----------GGAG---------TACAGGAGCTTT-----CT-------------CA--TC------------------------CGACCAGAC--C---------------------------------CGACCCCGAGGCC------------------------------------------------------------------------------------------------------TTGCGAGCATCTCCG--------------------TGATCCGC-ATCAGAGTG----------TCAA--TAACTAACCGGGTGACTCTGCACTACCCGTATCATTGCCTTGCT--TCCGCTG-C---------------------------CTTC-------CAAAA-----------G----AAAAGAAGGGAA-----ATCCTT--CAT----------------------------------------------------------------------------------------------------TTCCCGCTGGAATCC-------------------------------------------------CCT------------CA-----TTCGTGCAAC-CCTTTTCATGCTTGCCTTTC-----T------CTTAAT-TGCCTCCGACAGCTTC-TCTGGACCGCCCCAAGACGGAAATGTCACGGGCCGAGGCGGCCGCA | |
| droKik1 | scf7180000302476:160129-160454 - | CGTCTGCCACATGATAGCCTCGCCCACGGTCTCTCTGCAGAGCAACAAAGGTAAGCCCC----A---------AGGAGCACAGGGAGCACGAG-----------CG-------------AA--TC------------------------CGA---------------------------------------------------G---------------------------------------------------------------------------------------------------------------------------------A-------CCCCAATCCGTGAGCCAAGTG----------TAAATAAAACTAACCGGGTGAA---------CCCGTATCATTGCCTTGCT--TCCGCCG-CTGTGCCGTTGTTGTCGCCTCCTCCTGCCA-------------------------G----CAAAG------------ATT-CCACC--A------------------------C-------------------------------------------------------------------------CCTCCAGCAGCATT-C-------------------------------------------------CTG------------CTTTGT-CC--CGCAAC-CCGACTCATCTTT------------TATAATTTTCCAT-TGACTTCGACAGCTTC-ACTGGACCGGCCCAAGACGGAGATGTCGCGGGCGGAGGCCGCCGCC | |
| droFic1 | scf7180000453342:769737-770050 + | CGTCTGCCACATGATATCATCGCCCACGGTTTCGCTGCCGAGCAACAAAGGTAAGCCCGGGAAAAAG-TCGCGAGGAG---------CACGAG--GTCG-----CC-------------AG--CC-----------------------------------------------------------------------------------CGT-T-------------------------------------------------------------------------------------------------------------------------------TCTCGAGTCCGTGAGAAAAGTGTACTGTATGGTAGA--TAGCCAACCGGGCGAATGCGCAGCGCCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CTCC-------GAAAT-----------G----AAAAGAAAAG-------AATCCC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------C------------AATTGAATT--GGCCAA-CCCACTCACGTCTGCTTTT------C------CCCCAT-TCG-ACCGACAGCTTC-ACTGGACCGACCCAAAACCGAGATGTCGCGGGCCGAGGCCGCTGCT | |
| droEle1 | scf7180000491214:4010092-4010433 - | TGTCTGCCACATGATAGCATCGCCCACGGTCTCACTGCCGAGTCAGAAAGGTAAGCCAGGCCCAAAGGAAGCGAGGGT---------CATAAG--GTCA-----CG-------------AGGGTC--ATAAGGTCACGAAGGT------CA-----------------------------------------------TCTGAGGCC------------------------------------------------------------------------------------------------------CAGTGGG-----------TCATTCC-------CAAAAATCCGTGAGCAACGTG----------TTAA--TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CA-------------------------G----AAAAG------------ATTTCCCAC--A------------------------AA--ACA----------------------------------------------------------------------------------T-------------------------------------------------CTC------------CATTGAGTG--CACAAC-CCAACTCATGTCCCCTTT-------T------CCCCAT-TGC-ATCGACAGCTTC-GCTGGACCGTCCCAAAACGGAGATGTCCCGGGCCGAGGCAGCCGCC | |
| droRho1 | scf7180000776849:678770-679110 + | CGTCTGCCACATGATAGCATCGCCCACGGTGTCGCTGCCGAGCAACAAAGGTAAGCCAGGCTCAAGG-CCGCGTCGAG---------CACGAG--GTCA-----CG-------------AA--TC------------------------GAATTGGGT--CC----------------------------------------------CTT-TTCCGTCGCCCAGC-----------------------------------G----------------------------------------------------AG-----------GC---------GTTCTGAAATCCGTGAGCAAAGTG----------TAAA--TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CTCA-------AAAAA-----------T----AAAAG------------ATTTCCCCC--A------------------------CA--ACG----------------------------------------------------------------------------------T-------------------------------------------------TTC------------CATTGAGTT--TGCAGC-CCTACTCATGTTTGCTTT-------T------CCCCAT-TGA-ACCGACAGCTTC-ACTGGACCGCCCCAAAACAGAAATGTCCCGGGCGGAGGCTGCCGCC | |
| droBia1 | scf7180000302143:259880-260231 + | CGTCTGCCACATGATAGCATCGCCCACGGTGTCCCTGCCGAGCAACAAAGGTAAGCCAGG-------------AGGAG---------CACAAG--GTCA-----CG-------------AA--TC--A-AGAATCACGAATTTCGCATCGAATTGAGT--T------------------------------------ACCCAGTGCCCCGT-T--------------------------------------------------------------------------------------------------------------------------------TGAATATCCGTAAGCAAAGTG----------TAAA--TAACTAACCGGTTGAATGCGCAACGCCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CTCC-------AAAAG-----------G----AAGAG------------ATTCCC--CACA------------------------TCTGGTT------TATTTCCC-----------------------------------------------------------------------------------------------------------------------TT---------CCCCATTGAATT--CGCCGC-CCCACTCATGTTTGCCTTTC-----A------CCCCGT-TGA-ACCGACAGCTTC-ACTGGACCGTCCCAAAACGGAAATGTCGCGGGCCGAGGCTGCGGCT | |
| droTak1 | scf7180000415878:857122-857431 + | CGTCTGCCACATGATAGCATCGCCCACGGTCTCGCTGCCGAGCAACAAAGGTAAGCCG-------------CCAGGAG---------CACAT--------------------------------C------------------------GAACTGAGT--CCCAAGTT----A-------T--------------------------CCGT-G--------------------------------------------------------------------------------------------------------------------------------TCCAGAGCCGTAAGCACAGTG----------TAAA--TAACTAACCGGGTGAATGCGCAACGCCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CTCC-------GAAAG-----------G----AAAAG------------AATCCTCTC--C------------------------TC--C----------------------------------------------------------------------------------------------------------------------------------------T------------CTTTAAATT--CGTAGCCCCCACTCATGTTTGCTTTTTCCCTCC------CCCCGT-TGA-ACCGACAGCTTC-ACTGGACCGCCCCAAAACGGAAATGTCGCGGGCCGAGGCTGCGGCC | |
| droEug1 | scf7180000409672:10053-10380 - | CGTCTGCCACATGATAGCATCGCCGACAGTCTCCCTGCCAAGTAACAAAGGTAAGCCAGGCCCAAGGGCCGCGAGGAG---------CACAGG--GTCG-----CG-------------AA--TC------------------------C---------------------------------------------------------------------------------------------------------------------------------------------------------------ACG---------------------AACTCGTTCTTACATCCGTGAGCAAAGTG----------TATA--TAACTAACCGGGTGAAAGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------ACCC-------AAAAA----------AG----AAAAT------------ATTCTT--CACA------------------------TACGGTTTTTCTTTTT-----TATCTTTTTTGAA----------------------------------------------------------------------------------------------------ATCTTAC-----------------------------------TCAATGCTTGATTCT------T------CCCAAT-TGA-ACCGATAGCTTC-CTTGGACCGCCCCAAAACGGAAATGTCACGGGCCGAAGCTGCGGCT | |
| dm3 | chr2R:11152432-11152777 - | CGTCTGCCACATGATAGCATCGCCTACGGTATCCCTGCCAAGTAATAAAGGTAAGCCAGGCCCAAGG-GCTCGAGGAG---------CACATG--GTTA-----CG-------------AA--TC------------------------CGAATGAGC--CC-TAGTC----A-------T--------------------------CCGT-TTTTGCTTACCATC-----------------------------------G----------------------------------------------------AG-----------GC---------ATTCCTACATCCGTGAGCAAAGTG----------TG----TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CACC-------AAAGC-----------G----AAATG------------ATTTCCATT--A------------------------CATGATT----------------------------------------------------------------------------------T-------------------------------------------------TTG------------TATTGAATT--TGAAAC-CGTACTCACCCTTATATT-------A------A----CTTTA-ACTGACAGCATC-TTTGGACCGCCCCAAAACCGAAATGTCGCGGGCTGAGGCTGCGGCC | |
| droSim2 | 2r:11822321-11822669 - | dsi_22914 | CGTCTGCCACATGATAGCATCGCCTACGGTATCCCTGCCAAGTAACAAAGGTAAGCCAGGCCCAAGG-GCGCGAGGAG---------CACATG--GTTA-----CG-------------AA--TC------------------------CGAATGAGC--CC-TAGTA----A-------C--------------------------CCGT-TTTTGCTTACCATC-----------------------------------G----------------------------------------------------AG-----------GT---------ATTCCTACATCCGTGAGCAAAGTG----------TG----TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CACC-------AAAGC-----------G----AAATG------------ATTTCCATC--A------------------------CATGATT----------------------------------------------------------------------------------T-------------------------------------------------TTG------------TATTGAATT--TGAAAC-CGTACTCACCCTTATATT-------A------CCTTAT-TTA-AATGACAGCATC-TTTGGACCGCCCTAAAACCGAAATGTCGCGGGCTGAGGCTGCGGCT |
| droSec2 | scaffold_1:8645081-8645429 - | CGTCTGCCACATGATAGCATCGCCTACGGTATCCCTGCCAAGTAACAAAGGTAAGCCAGGCCCAAAG-GCGCGAGGAG---------CACATG--GTTA-----CG-------------AA--TT------------------------CGAATGAGC--CC-TAGTC----A-------C--------------------------CCGT-TTTTGCTTACCATC-----------------------------------G----------------------------------------------------AG-----------GT---------ATTCCTACATCTGTGAGCAAAGTG----------TG----TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CACC-------AAAGC-----------G----AAATG------------ATTTCCATC--A------------------------CATGATT----------------------------------------------------------------------------------T-------------------------------------------------TTG------------TATTGAATT--TGAAAC-CGTACTCACCCTTATATT-------A------CCTTAT-TTA-AATGACAGCATC-TTTGGACCGCCCTAAAACCGAAATGTCGCGGGCTGAGGCTGCGGCT | |
| droYak3 | 2R:11074100-11074456 - | CGTCTGCCACATGATTGCATCGCCCACGGTATCCCTGCCAAGTAACAAAGGTAAGCCAGGCCCAAAAGCCGCGAGGAG---------CACAAG--GTTATCCAATG-------------AA--TC------------------------CGAATGAGC--CC-TAGTC----A-------C--------------------------CCGTTTTTTGCTTACCATC-----------------------------------G----------------------------------------------------AG-----------GC---------GTTTCAACAACCGTGAGCAAAGTG----------TG----TAACTAACCGGGTGAATGCGCAATACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CACC-------AAAGC-----------G----AAATG------------AATGCCATCACA------------------------CACGATT----------------------------------------------------------------------------------T-------------------------------------------------T-T------------TATTGAATT--TGCAAC-CCAACTCACGCTTGTTTT-------A------CCCCAT-TTA-AATGACAGCTTC-TTTGGACCGGCCCAAAACAGAAATGTCGCGGGCTGAAGCTGCGGCT | |
| droEre2 | scaffold_4845:14717634-14717981 + | CGTCTGCCACATGATAGCATCGCCCACGGTATCCCTGCCAAGCAACAAAGGTAAGCCAGGCCCGAGG-CCGCGAGGAG---------CACATG--GTTA-----CA-------------AA--TA------------------------CAAACGAGC--CC-CAGTC----A-------C--------------------------CCGT-TTTTGCTTACCATC-----------------------------------G----------------------------------------------------AG-----------GC---------GTTTCAACATACGTGAGCAAAGTG----------TG----TAACTAACCGGGTGAATGCGCAACACCCGTATCATTGCCTTGCT--TCCGCCG-C---------------------------CACC-------AAAGC-----------G----AAATG------------AATCCCATC--A------------------------CATGATT----------------------------------------------------------------------------------T-------------------------------------------------T-T------------TATTGAATT--TGCAAC-CCTACTCACGCCCATTTC-------A------CCCCAT-TTA-AATGACAGCTTC-CTTGGACCGCCCCAAAACAGAAATGTCGCGGGCTGAGGCTGCGGCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 10:06 PM