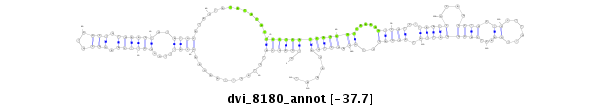
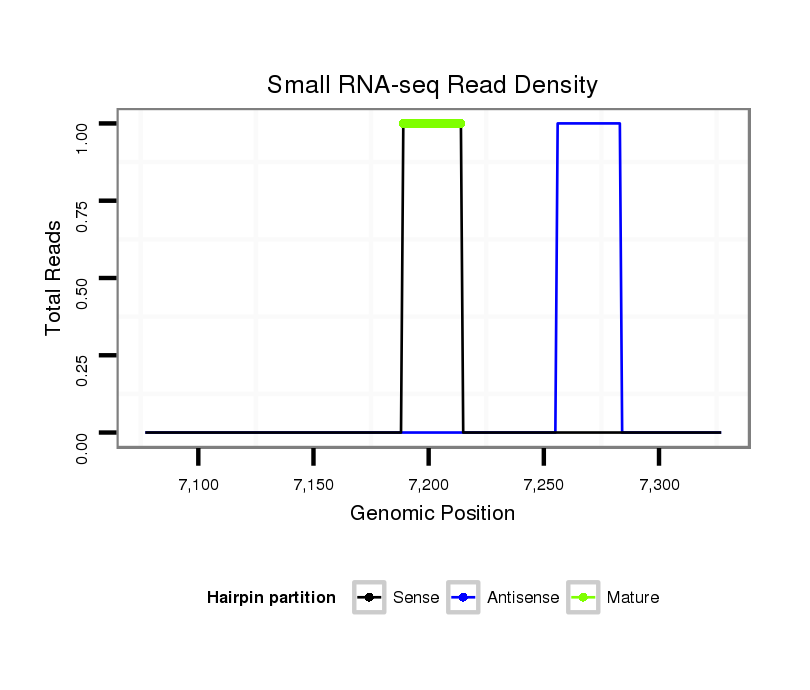
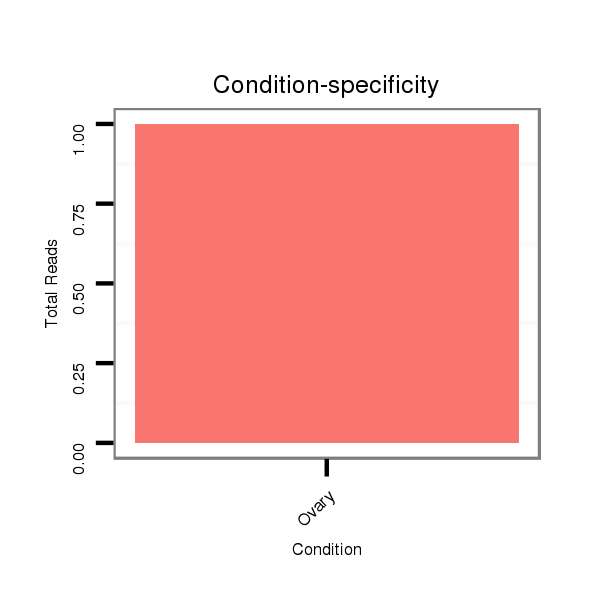
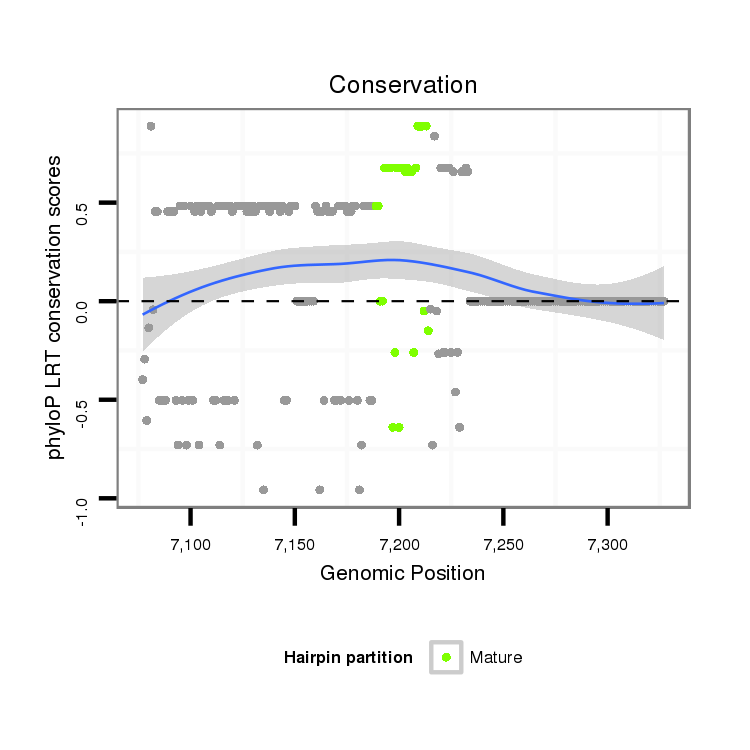
ID:dvi_8180 |
Coordinate:scaffold_12928:7127-7277 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.7 | -37.6 | -37.6 |
 |
 |
 |
CDS [Dvir\GJ16700-cds]; exon [dvir_GLEANR_17203:1]; intron [Dvir\GJ16700-in]
No Repeatable elements found
| -------------------------------###################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATCTGGGTCGGGGGCACAAAGCACCATCAAATGTGTGAAATACCAAATTGTGAGCATCTACTTAAGAAGCATTTGGGGTTCAAGGCGTGGCTGTGAACTGTATGCATACTATATATATTGCTCATCAGCGTTAAAAGTCGTCTTAATCGCAGCCTGCTCCTTGTTGAACTCAGGGATTATACGACCTGCAGCTGACAAATTTGTTATGTGAGTTACGTTTGCCGAAAGCAGAAGGCATAACTTACAAGTT **************************************************.((((((............((((....((((((.(((...))).))))))..)))).............))))))(((((((.....(((((..(((((....((((.((......))...))))))))).)))))..)).))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
V047 embryo |
V116 male body |
SRR060657 140_testes_total |
SRR060666 160_males_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................TATATATTGCTCATCAGCGTTAAAAG................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TGCATACTCTATATCTTGCTGA.............................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GCTCCTAGTTGAACTTTGGG........................................................................... | 20 | 3 | 7 | 0.29 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................GCTCGTAGTTGAACTCTGGG........................................................................... | 20 | 3 | 8 | 0.25 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................GTTCAAGTCCTGGCGGTGA.......................................................................................................................................................... | 19 | 3 | 17 | 0.18 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................TTTTTCAGAAGCATTTGGGG............................................................................................................................................................................ | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................AGCATTCTATACATATTGCT................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GTAGACCCAGCCCCCGTGTTTCGTGGTAGTTTACACACTTTATGGTTTAACACTCGTAGATGAATTCTTCGTAAACCCCAAGTTCCGCACCGACACTTGACATACGTATGATATATATAACGAGTAGTCGCAATTTTCAGCAGAATTAGCGTCGGACGAGGAACAACTTGAGTCCCTAATATGCTGGACGTCGACTGTTTAAACAATACACTCAATGCAAACGGCTTTCGTCTTCCGTATTGAATGTTCAA
**************************************************.((((((............((((....((((((.(((...))).))))))..)))).............))))))(((((((.....(((((..(((((....((((.((......))...))))))))).)))))..)).))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060684 140x9_0-2h_embryos_total |
M047 female body |
SRR060687 9_0-2h_embryos_total |
V053 head |
SRR060677 Argx9_ovaries_total |
SRR060664 9_males_carcasses_total |
M027 male body |
SRR060672 9x160_females_carcasses_total |
V116 male body |
SRR060666 160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
V047 embryo |
M061 embryo |
SRR060667 160_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060668 160x9_males_carcasses_total |
SRR060658 140_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........CTCCCGTGTTTTGTGGTAC.............................................................................................................................................................................................................................. | 19 | 3 | 19 | 7.37 | 140 | 36 | 19 | 11 | 1 | 0 | 6 | 4 | 5 | 1 | 8 | 10 | 3 | 8 | 0 | 1 | 7 | 6 | 1 | 4 | 0 | 3 | 3 | 2 | 0 | 1 |
| ..........CTCCCGTGTTTTGTGGTA............................................................................................................................................................................................................................... | 18 | 2 | 5 | 2.60 | 13 | 2 | 3 | 0 | 0 | 0 | 2 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TTCGTCTACGGTATTGGATG..... | 20 | 3 | 12 | 1.50 | 18 | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................TATGCTGGACGTCGACTGTTTAAACAAT............................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCTCCCGTGTTTCGTGGTA............................................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCCCCCGTGTTTTGTGGTAC.............................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.67 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAGTCGAAACTTTCAGCAGAGT......................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAGTCGAAAGTTTCAGCAGAGT......................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........GCTCCCGTGTTTTGTGGTAC.............................................................................................................................................................................................................................. | 20 | 3 | 5 | 0.40 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CGTCCCCCGTGTTTTGTGGTA............................................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAGTCGAAATTTTAACCAGAAT......................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGTCGCAAATTTCACCAGAGT......................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TCTCCCGTGTTTCGTGGTAC.............................................................................................................................................................................................................................. | 20 | 3 | 14 | 0.21 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CCTCCCGTGTTTTGTGGTA............................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GCTTTCGTCTACGGTATTGG........ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CTTCGTATACCACAGGTTCC..................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CCCCCGTGTTTTGTGGTACA............................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TTTCGTCTACGGTATTGGATG..... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TATGAGATGTATCACGAGTAG............................................................................................................................ | 21 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CCCACCGTGTTTTGTGGTA............................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGCCTCCGTGTTTTGTGGT................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12928:7077-7327 + | dvi_8180 | CATCTGGGTCG------------GGGGCACAAAGCACCATCAAATGTGTGAAAT---------ACCAAATTGTGAGCATCTACTTAAGAAGCATTTGGGGTTCAAGGCGTGGCTGTGAACTGTATGCAT--------------ACTATATATATTGCTCATCAGCGTTAAAAGTCGTCTTAATCGCAGCCTGCTCCTTGTTGAACTCAGGGATTATACGACCTGCAGCTGACAAATTTGTTATGTGAGTTACGTTTGCCGAAAGCAGAAGGCATAACTTACAAGTT |
| droMoj3 | scaffold_6359:1337530-1337680 + | TGGCTGGGCTATCTAGCAAGGTCAGGGCGAAGATTATCAGCAAATGCATTAGACCCAATCGCCACTAAATTGTGAGAATGTACTTAAGAGACATT---------AGCCATGGCCATAAACCGTACCAATTTACATATCCCAATATCATA------------------TAAAAATGGTC------------------------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_72:7126-7172 - | TATATA-------------------------------------------------------------------------------------------------------------------------------------------------TATTCTTGATCAGCATTAATAGCCGAGTCGATTGAGCCCTG---------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/16/2015 at 08:18 PM