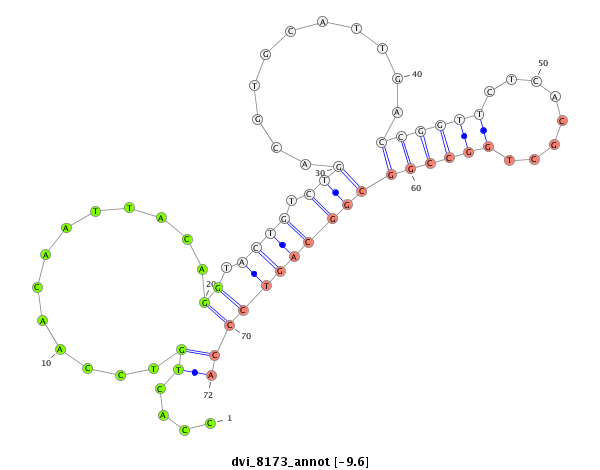
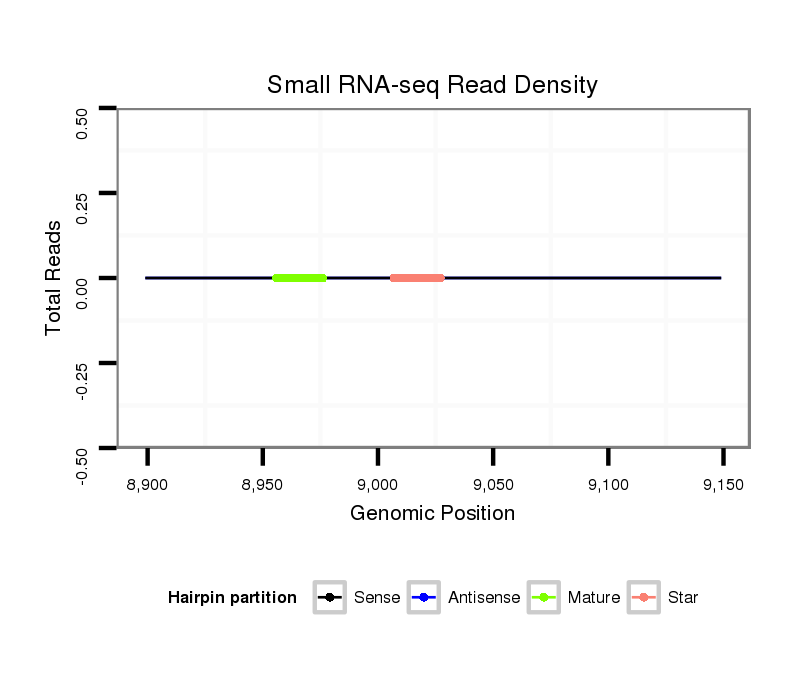
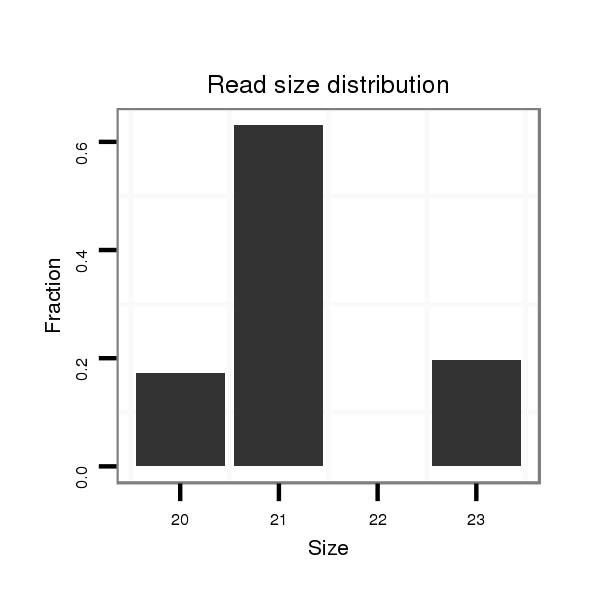
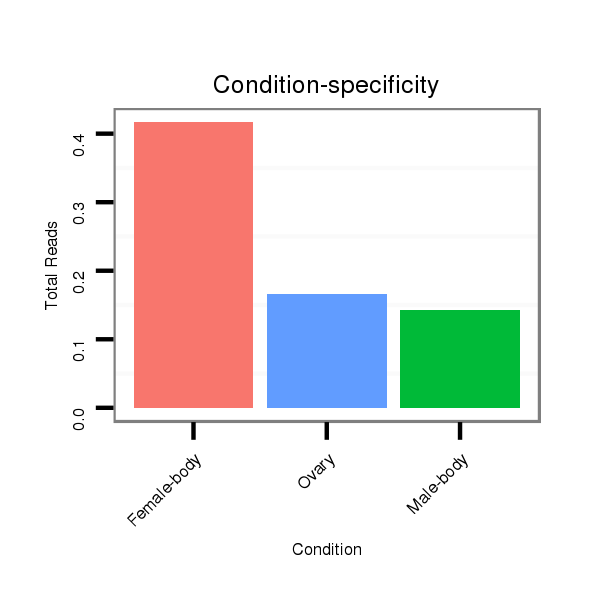
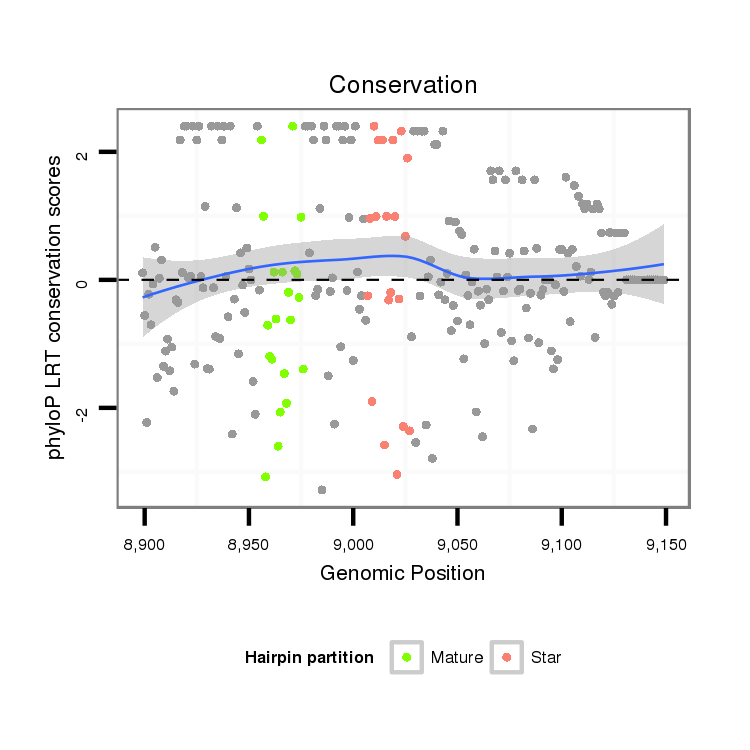
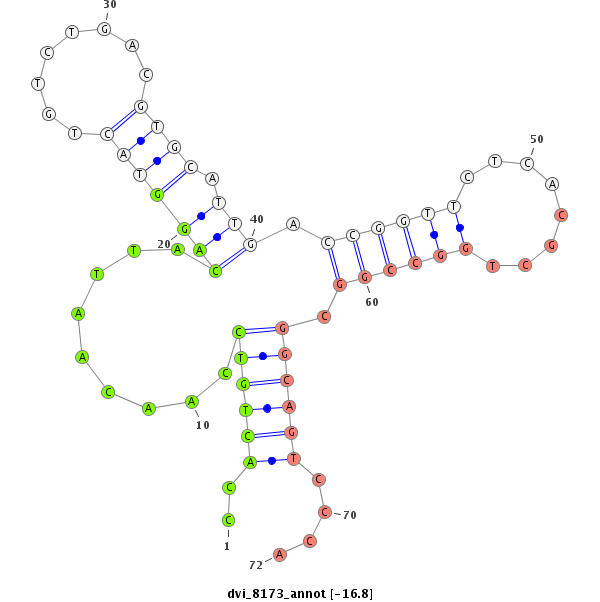
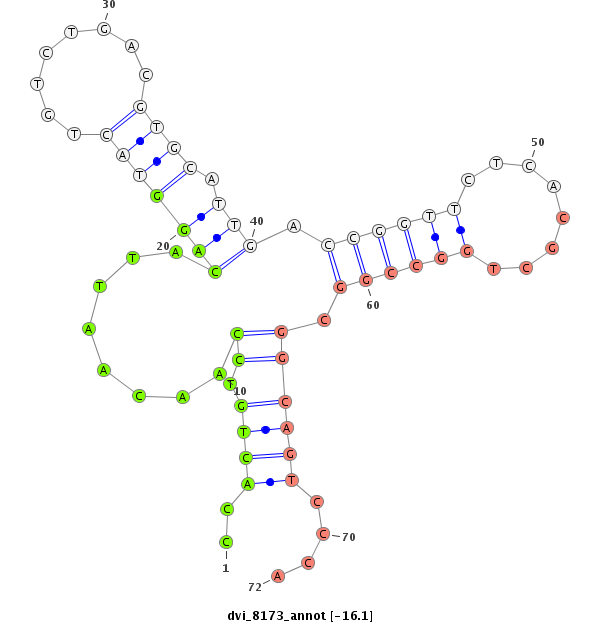
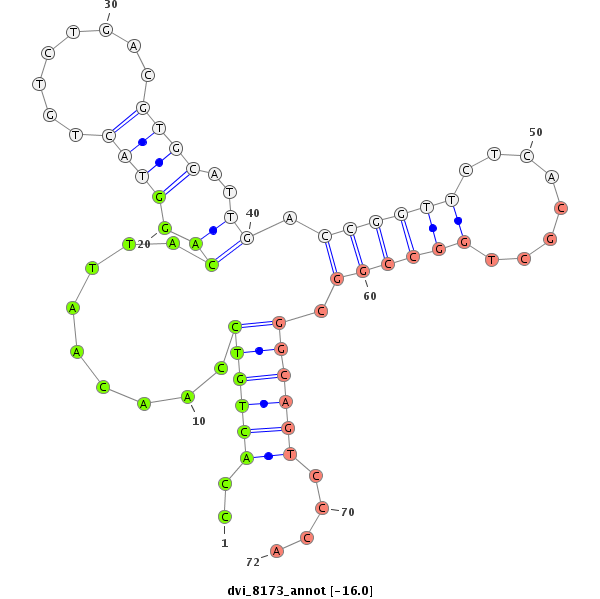
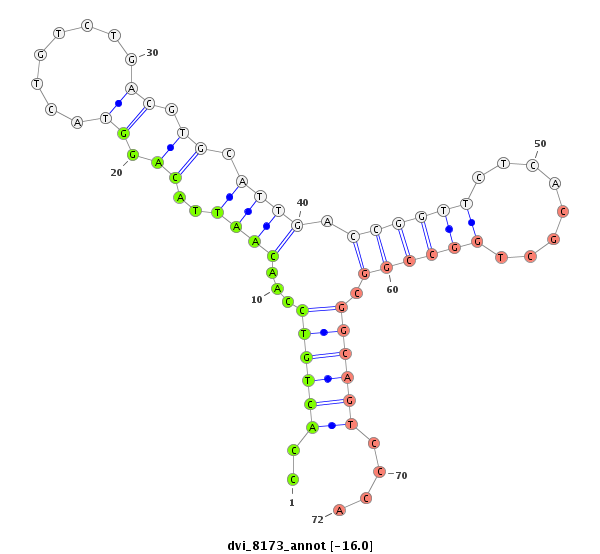
ID:dvi_8173 |
Coordinate:scaffold_12878:8949-9099 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.8 | -16.1 | -16.0 | -16.0 |
 |
 |
 |
 |
exon [dvir_GLEANR_17579:2]; CDS [Dvir\GJ17076-cds]; intron [Dvir\GJ17076-in]
| Name | Class | Family | Strand |
| Gypsy-4_DTa-I-int | LTR | Gypsy | + |
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCAACCTGCCTAAAAAGCGATTTGAACATATCCATATTGATTTAATTGGCCCTGTTGCCACTGTCCAACAATTACAGGTACTGTCTGACGTGCATTGACCGGTTCTCACGCTGGCCGGCGGCAGTCCCAATAACATAACGGCAGAGACGGAAGCTAAATTGCATGTTTGGATTGCAAACTACGGCATGCCACACTTCACAGCTACCGCCGCTCAAAGCACCTTATGAGGGGCCATTTCTAGTTATCAGCCG *********************************************************....((.............((.((((.(((...........((((((........)))))))))))))))))************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
M047 female body |
M027 male body |
SRR060675 140x9_ovaries_total |
SRR060670 9_testes_total |
SRR060689 160x9_testes_total |
SRR060667 160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................TTGGCCCTGTTGTCACTGTA.......................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCACTGTCCAACAATTACAGG............................................................................................................................................................................. | 21 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CGCTGGCCGGCGGCAGTCCCA.......................................................................................................................... | 21 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................ATTACAGGTACTGTCTGACGTGC.............................................................................................................................................................. | 23 | 0 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TTGGATTGCAAACTACGGCATC............................................................... | 22 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................TTGGATTGCAAACTACGGCCTC............................................................... | 22 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................ATTACAGGTACTGTCTGACGTGCATC........................................................................................................................................................... | 26 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TACAGGTACTGTCTGACGTGC.............................................................................................................................................................. | 21 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TTACAGGTACTGTCTGACGT................................................................................................................................................................ | 20 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TACTGTCTGACGTGCATTGACCC...................................................................................................................................................... | 23 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................ACAGGTACTGTCTGACGTGCATC........................................................................................................................................................... | 23 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................AGGTACTGTCTGACTTGCATTGACC....................................................................................................................................................... | 25 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TCTTTGGCCCTGTTGTCACTG............................................................................................................................................................................................ | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AGTTGGACGGATTTTTCGCTAAACTTGTATAGGTATAACTAAATTAACCGGGACAACGGTGACAGGTTGTTAATGTCCATGACAGACTGCACGTAACTGGCCAAGAGTGCGACCGGCCGCCGTCAGGGTTATTGTATTGCCGTCTCTGCCTTCGATTTAACGTACAAACCTAACGTTTGATGCCGTACGGTGTGAAGTGTCGATGGCGGCGAGTTTCGTGGAATACTCCCCGGTAAAGATCAATAGTCGGC
**************************************************************************************************************************....((.............((.((((.(((...........((((((........)))))))))))))))))********************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
V047 embryo |
V053 head |
M027 male body |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
SRR060675 140x9_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
GSM1528803 follicle cells |
SRR060657 140_testes_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........GATTTTTCCCTAAACTTG................................................................................................................................................................................................................................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GTGGAATACTCCCCGGTAAAGATCAA........ | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TTCGTTGAATACTTCACGGTA................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GGAATACTCCCCGGTAAAGATCAATAGT.... | 28 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................ACGGTGACAGGTTGTTAATGTCCATGAC........................................................................................................................................................................ | 28 | 0 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TTGCCACCTCTGCCTTCG................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GTGAAGTGGTGATGGCGG.......................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................CGGTGACAGGTTGTTAATGTCCATGAC........................................................................................................................................................................ | 27 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CAACGGTGACAGGTTGTTAAT................................................................................................................................................................................. | 21 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GGTGACAGGTTGTTAATGTCCATGAC........................................................................................................................................................................ | 26 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GGTGACAGGTTGTTAATGTCCATGACA....................................................................................................................................................................... | 27 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................GTTTGATGCCGTACGGTGTGA........................................................ | 21 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................TCCATGACAGACTGCACGTAACT......................................................................................................................................................... | 23 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GACAGGTTGTTAATGTCCAT........................................................................................................................................................................... | 20 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AAGTTGTTAAAGTGCATGACA....................................................................................................................................................................... | 21 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ...............................................................AGGTTGTTAAAGTGCATGA......................................................................................................................................................................... | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................GTTGTTAATGTGCATGAAA....................................................................................................................................................................... | 19 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................TTGTTAATGTGCATGAAA....................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......CAGAATTTTCGCTAAACATG................................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TCAACGGGGAAAGGTTGTTA................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CCGTCCCTGCCTTAGA................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................CGGCGAGATTCGTGGTAA........................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12878:8899-9149 + | dvi_8173 | TCAACCTGCCTAAAAAGCGATTTGAACATATCCATATTGATTTAATTGGCCCTGTTGCCACTGTCCAACAATTACAGGTACTGTCTGACGTGCATTGACCGGTTCTCACGCTGGCCGGCGGCAGTCCCAATA----ACATAACGGCAGAGACGGAAGCTAAATTGCATG------TTTGGATTGCAAACTACGGCATGCCACACTTCACAGCTACCGCCGCTCAAAGCACCTTATGAGGGGCCATTTCTAGTTATCAGCCG |
| droMoj3 | scaffold_6541:89127-89279 + | ACGCACTCACATCTCAACGATTCGAACATATTAACTTGGATCTGATTGGACC-ACTACCTCAATCAGGACAATACAGATACTGTCTCACTATCATCGATCGCTACACCAAGTGGCCAGAAGCCGTACCTATCGAAGATATACAAGCAGAGACAG----------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004569:22038-22190 + | TACCCGTTCCGGATAACAGATTCGACCACGTGCACTTGGACATCATAGT----AATGCCTTACTATCAAAACTTCCGCTACTGCCTAACGATGATCGACCGATTTTCACGTTGGCCCGAAGCTGTC--TCTCACTGACATCACTGCAGAGACAATTGCA------------------------------------------------------------------------------------------------------ | |
| dp5 | Unknown_group_2:24856-25003 + | TCACCGTCCCCGATGGCAGGTTTGATCACATACATCTAGACATTGTTG---T-GATGCCCTTTGTTGACGGATACAAATATTGTCTGACAATGATTGACCGTTTTTCCCGCTGGCCGGAGGCTGTGCCGCTGAAGGATATGACTGCAGAGAC------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_59:139997-140219 + | TCACCGTCCCCGATGACAGGTTTGATCACATACATCTAGACATTGGT---GT-GATGCCCTTTGTTGACGGATACAAATATTGTCAGACAATGATTGACCGTTTTTCCCGCTGGCCGGAGGCGGTGCCTCTGAAGGATATGACTGCAGAGACCATATGTGAAGCAATATGGTCGCGATGAATATCACGTTTTGAATGTCCCTT---GACTATTACAACCGATCAAGGTAC------------------------------- | |
| droAna3 | scaffold_13756:33069-33282 - | ATGCCGCCAGCGACGCCCGTTTCGAACACATCAATATCGACATCGTTGGCCC-ACTGCCATCATCGAACGGATACCGATACTGCCTCACGTGTATCGATCGTTTCTCAAGATGGCCCGCAGCAATCCCTTTAGTCGACATCACAGCACAATCTGTGGCCAACGCTCTTATTTCGGGCTGGTTCGCTCTGTACGGTATCCCTCT---CGACATCACAAC------------------------------------------- | |
| droBip1 | scf7180000393686:15697-15912 - | ACACCGCCAGCGACGCCCGTTTCGAACACATAAATATCGACATAGTCGGCCC-GCTGCCTTCATCGAACGGATACCGATACTGCCTCACGTGTATCGATCGTTTCTCAAGATGGCCCGCAGCAATTCCTTTAGTAGACATCACAGCACAATCTTTAGCTAACGCACTGATAACGGGCTGGTTCGCTCAGTACGGTATTCCTCT---CCACATCACAACCG----------------------------------------- | |
| droKik1 | scf7180000299059:7877-8033 + | TCACGCTTCCTACTCAAAGATTTGACCACATCAATCTGGATCTCATCGGACC-ATTACCCCAATCAGGTCAATATAGATACTGCTTAACAATCATTGACCGTTATACACGTTGGCCGGAAGCCATCGCAATCGAGGACATGCACGCCAACACGGTAGC------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453919:4549-4762 + | ATGTCGCCAGCGACGCCCGTTTCGAACACATCAACATCGACATCGTTGGCCC-ACTGCCATCTTCGAACGGATACCGATACTGCTTGACGTGTATCGATCGCTTCTCAAGATGGCCCGCAGCAATTCCCTTAGTCGACATCACAGCACAATCTGTAGCCAACCCGCTGATTTCGGGCTGGTTCGCTCAGTACGGAATCCTTCT---CCACATCACAAC------------------------------------------- | |
| droEle1 | scf7180000489069:48481-48638 + | TCACAGTTCCCAGCGACAGATTCAACCATGTTCATCTGGACATCGTCAT----CATGCCGTTCCATCAAAACTTTCGGTACTGCCTTACCATGATAGACAGATTTACTCGCTGGCCGGAGGCTGTCCCTCTTACAAACATATCGGCTGATACGGTGGCTAAA--------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000766317:1880-2078 + | TCGACGTGCCCGATGCCCGATTCCAACACATCAATATAGACCTGATCGGCCC-GCTACCACCGTCCAATGGTTACCGCTACTGTTTGACGGTCATCGACCGATTCAGCAGATGGCCAGCTGTATTACCAGTGACTGAGATGACTGCCGAAGAGGTGGCACTCGCACGCCTGCACGGATGGATCGCACATTATGGAGTGCC------------------------------------------------------------- | |
| droBia1 | scf7180000302398:356-476 - | TTACGCTTCCTTCACAAAGATTCCACCACATCGATCTGGACCTCATCGGACC-ATTACCCCAATCAGGTCAATATAGATACTGTTTAACAATCATTGACCGTTATACACGTTAGCCAGAAGC------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000412786:1868-2102 + | TACACGTTCCAAGCGGAAGATTCGATCATGTACATCTGGACATAGTAA---T-AATGCCAGCATATCAGAATTTCCGCTACTGTCTCACAATGATCGATCGCTTCTCACGTTGGCCTGAAGCTATACCACTAACCAACATTACCGCAGAAACGATTGCCACCGCCTTTTGGACCCACTGGATTTCACGATTTGGCGCCCCAAA---GACGATTACTACTGATCAGGGAACCCAGTTTGAGGC------------------- | |
| droYak3 | v2_chrUn_006:82007-82229 - | TCGCTGTGCCAAAATAAAGATTCGACCACGACCACCTGGATATAGTTGT----CATGCTTTCACAAAATAAATACCGCTATTGTCTTACCATGATTGACCGTTTCTCGCGATGGCCGAAAGCAGTACCACTCACTCATATCACGGCCGATACGATAGCCATGGCGTGCTGAACACACTGGATATCACGTTTTGGATCACCCAA---AACAATCATCACTGATCAAGGCAC------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/16/2015 at 08:25 PM