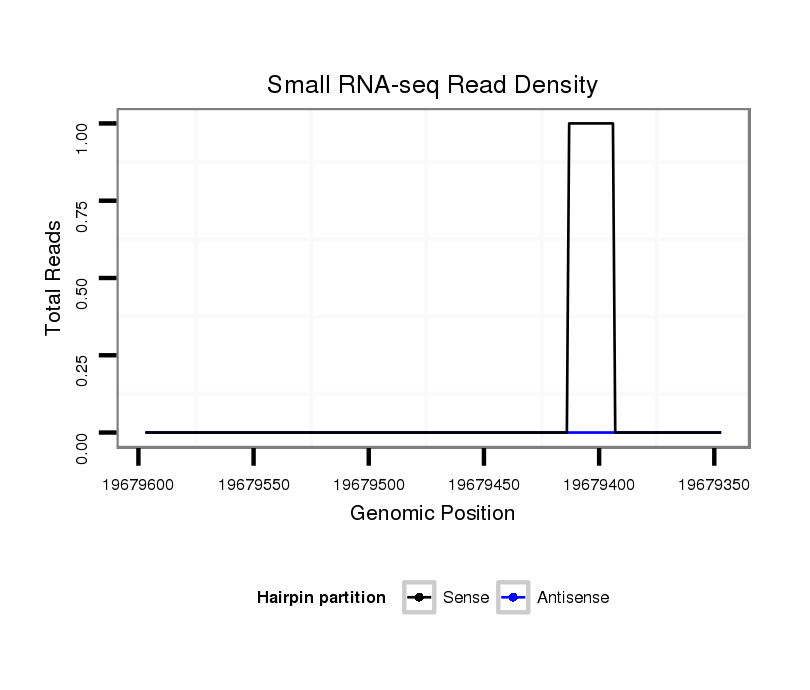
ID:dvi_8002 |
Coordinate:scaffold_12875:19679397-19679547 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ19918-cds]; exon [dvir_GLEANR_5388:2]; intron [Dvir\GJ19918-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGGGCGGCTGCTGACGGGGTTGTCAGGGGGGTTTATATGGGTTGGTGGGGAAGGGGAGGTTGGCTATTGTCAATAGCCTACAAATTGGCGCAGCGTACGGCTGCAATACATTTTTGAGTACCTCCACACATATTCTATAGCCACGGGGGTTGAGTAATCGACAACGCCAGCTATCGAACAATTCAGCGCCATATCGCACAGCTCCATTGCCATCGAGTGTGCCCAGGTGTGCAGAGCTGACGCAGCGGCAG **************************************************....((((((((((....)))))..(((........)))(((((.....)))))..(((........)))))))).......(((.(((((...((..(((((....)))))....)).))))).))).......(((......)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
V053 head |
SRR060666 160_males_carcasses_total |
V116 male body |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060675 140x9_ovaries_total |
SRR060668 160x9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................AGCGCCATATCGCACAGCTC............................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ATGCCATCGGGTGTGCCCAGAT....................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TGGCAGGGGGGCTGATATGGGT................................................................................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTCAAGCCACGCGGGTTGAG................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GCTGCTTACGGTGTCGTCA.................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| ..............................................................................................................................................................................................................TTGCCATCGGATGTGCCCAGAT....................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GGGAAGGTGAGGTCGGCGATT....................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TGGCAGGGGGGTTGATAGGGG.................................................................................................................................................................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................TGGCAGGGGGGTTGATAAGG................................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GGCCGCTGCTGACTGGGT....................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGGTGGAGAGGGGGAGGT............................................................................................................................................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................GGAGGGGAAGGGGAG................................................................................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TGGCGGGGTTGTTAGCGGG............................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACCCGCCGACGACTGCCCCAACAGTCCCCCCAAATATACCCAACCACCCCTTCCCCTCCAACCGATAACAGTTATCGGATGTTTAACCGCGTCGCATGCCGACGTTATGTAAAAACTCATGGAGGTGTGTATAAGATATCGGTGCCCCCAACTCATTAGCTGTTGCGGTCGATAGCTTGTTAAGTCGCGGTATAGCGTGTCGAGGTAACGGTAGCTCACACGGGTCCACACGTCTCGACTGCGTCGCCGTC
**************************************************....((((((((((....)))))..(((........)))(((((.....)))))..(((........)))))))).......(((.(((((...((..(((((....)))))....)).))))).))).......(((......)))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060669 160x9_females_carcasses_total |
SRR060675 140x9_ovaries_total |
SRR060686 Argx9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................TTGCGGTAGATAGCCTGTTA..................................................................... | 20 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ..............................................................................................................AAAAACTCATTGCGGTATGT......................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ...CACCCACGACTGCCCCAGC..................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:19679347-19679597 - | dvi_8002 | TGGGCGGC---------TGCTG--------ACGGGGTT----GTCAGGGGGGTTTATATGGGTT-GGTGGGGAAGGG--------------GAGGTTG---GCTATTGTCAATAGCCTACAAATTGGCGCAGCGTACGGCTGCAATACATTTTTGAGTACCT--------------------------------------------------CC-ACACATAT-TCTATAGCCA-C-GGGGGTTGAGTAATCGACAACGC---CA---------------------GCT-------ATCGAACAATTCAGCGCCATATCGCA--CAGCTCCATTGCCATCGAGT-----GTGCCCAGGTGTGCAGAGCTGACGCAGC------GGCAG------------ |
| droMoj3 | scaffold_6496:6744394-6744667 - | TTTGTTGC---------TTTTG--------GCAGGGCTGGGGTGCAA---------TTTGGGTTAAGTGGGGGAGGGCATAGCGGTGCTGCGGGGTTA---GCCATTGTCAATAGCCTACAAATTGGCGCAGCGCATGGCGGCAATACATTTTTGAGTGGCT--------------------------------------------------CC-AATCGTATTTTTGTAGCCA-C-AGGGGTTGAGTAATCGACAAC--GGCAA---------------------GCT-------ATCCAACAATTCAGCGCCATATCGCACCGAGCTCCATTGCCATCGAGT-----GTGCCCAGGTGTGCAGAGCTGACGCAGCAG------CAGCAGTGGCAG--- | |
| droGri2 | scaffold_15245:17888766-17889034 + | GAGGGGGCCAATCTAGTTGCTGTTATGGCCATGGGGTT----GTAAGG----------------------GGAAGGG--------------GGGATGGGAACTTATTGTCAATAGCCTACAAATTGGCGCAGCGTATGTTTGCAATACATATTTGAGTACTT--------------------------------------------------GC-GCTCGT---TTTGTAGCCA-C-GGGGTTTGAGTAATCGACAAC-----------------GACATCGACCAGGT-------ATCCAACAATTCAGCGTCATATCGACAATAGCTCCATTGCCATCGAGT-----GTGCCCAGGTGTGCAAAGCTGACGCAGCAG------CGGCAGCAGCAGTTG | |
| droWil2 | scf2_1100000004510:2400507-2400773 + | TGCACTGG---------AAAAG--------ACGGGG------GGCAGGGGGATTGAAGTAGACT-CGAG-----------------------------GGACTTTTTGTCAATTGCCTACAAATTGGCAC-----------GCTCAACATTTTTGGCTTCCACAACGTCACATATACACAGACCCATATCTACAGATATGTATATATATATTCCGATACATATATCTAAAGCCAACGGGGGGTTGAGTAATCGGCTATACGGCAGGCGCTCCTCCTCCATCTCCATCGCCATCGCCATCC---------------------ATCTATATCCATGGCCATTGAGT-----TGGCCCAG------------------------------------------- | |
| dp5 | 3:5892566-5892660 + | CG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGATTGAGTAATCGGTTATCC---AG-----GCTCTGGCATCGACCATGT-----------------------------------------CATTGCCAGCGATTGTTGGGTGCCCAGGTGTGCC--------GCCGCCGCCGCTGCAC------------ | |
| droPer2 | scaffold_2:6093951-6094042 + | CG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGATTGAGTAATCGGTTATCC---AG-----GCTCTGGCATCGACCATGT-----------------------------------------CATTGCCAGCGATTGTTGGGTGCCCAGGTGTGCC--------GCCGCTGCCG---CCG------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/17/2015 at 05:03 AM