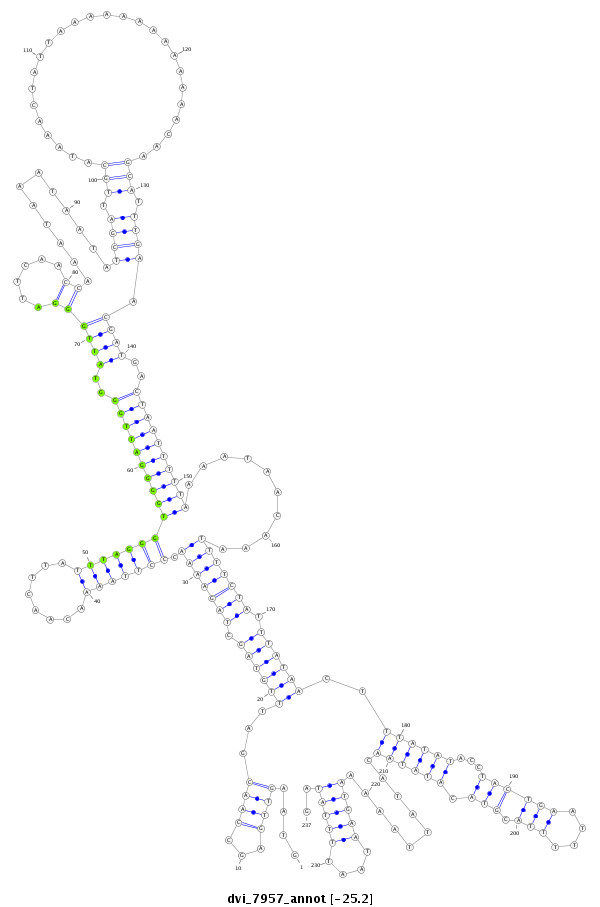
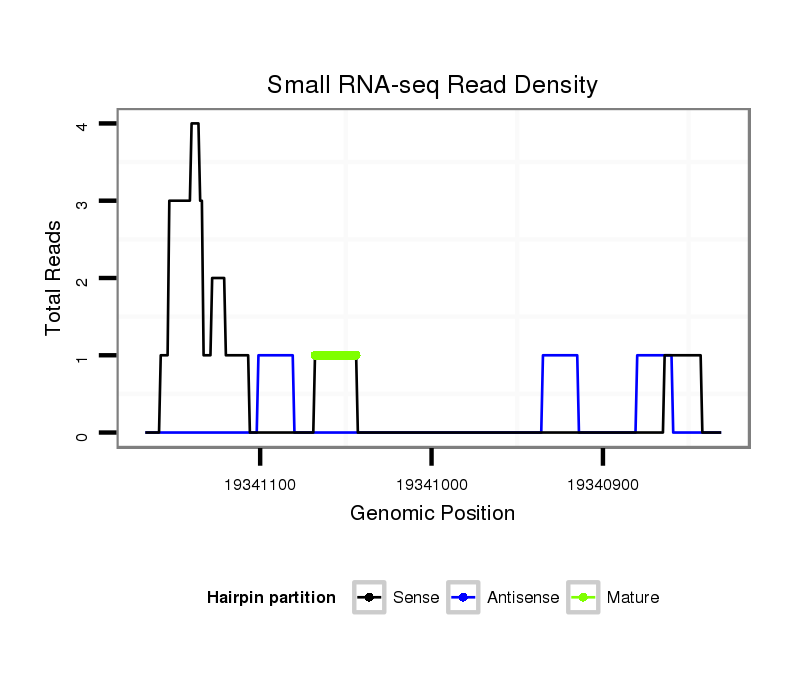
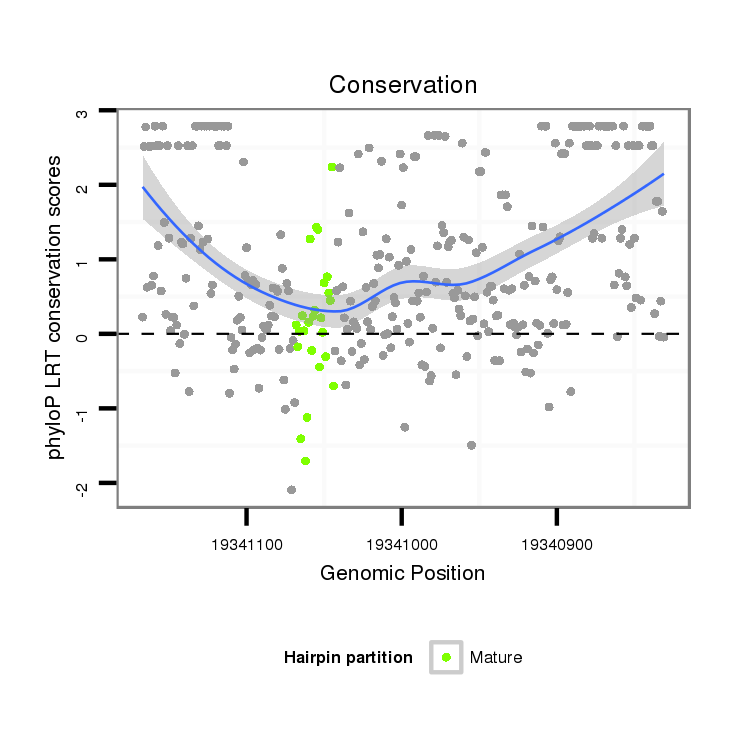
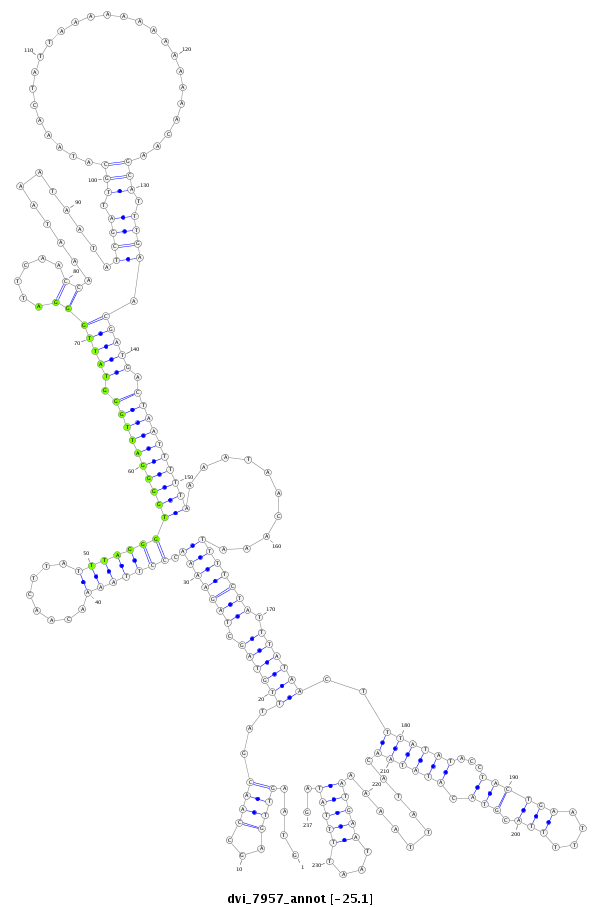
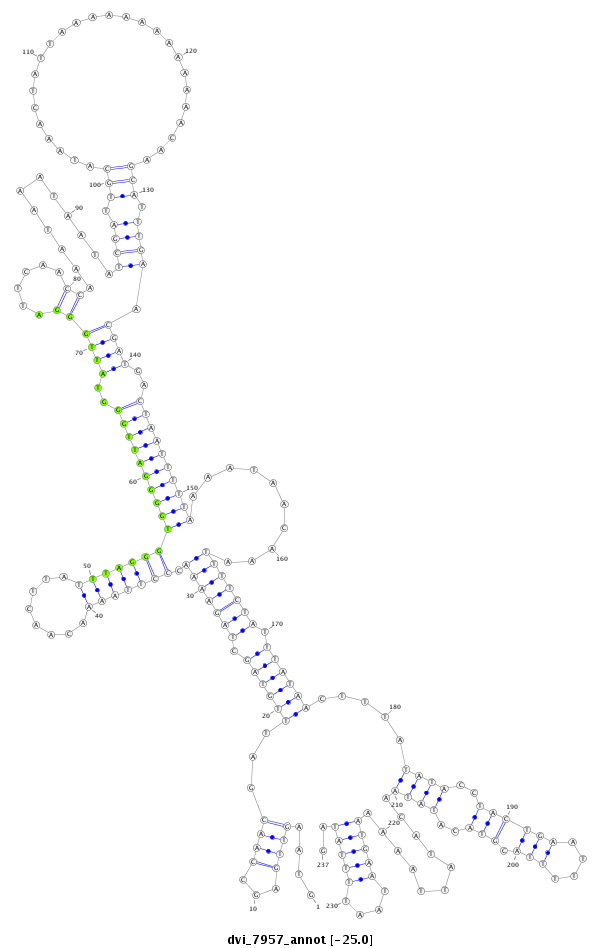
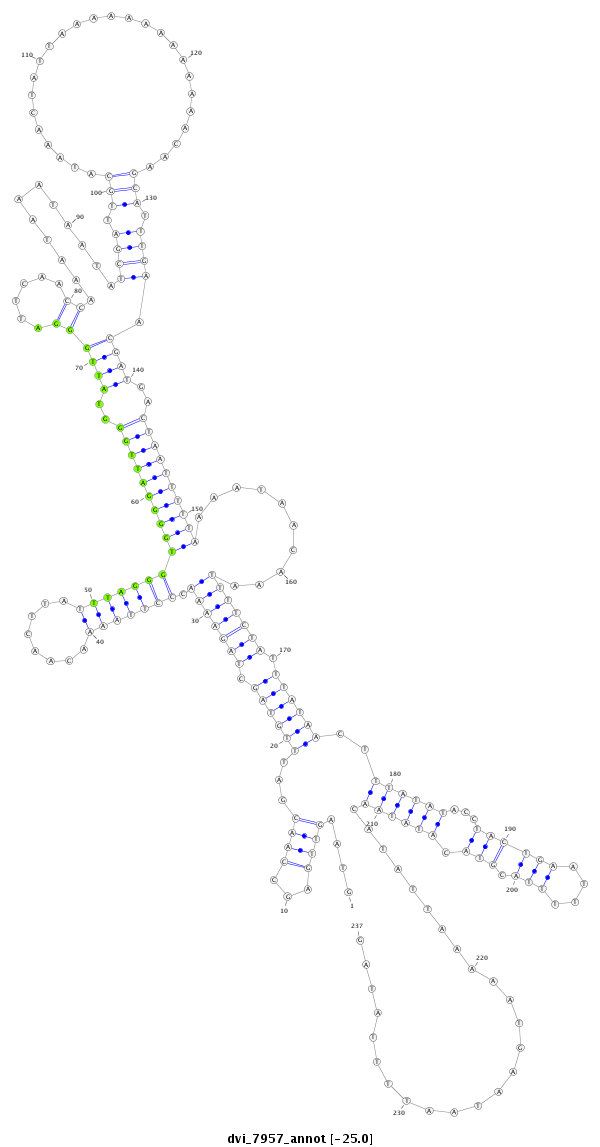
ID:dvi_7957 |
Coordinate:scaffold_12875:19340881-19341117 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -25.1 | -25.0 | -25.0 |
 |
 |
 |
CDS [Dvir\GJ19943-cds]; CDS [Dvir\GJ19943-cds]; exon [dvir_GLEANR_5410:6]; exon [dvir_GLEANR_5410:5]; intron [Dvir\GJ19943-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCATCGCATGCCCAAGCGTAGCGCACCATCGGGCTATGAATACACCAAACGTAAGTTGAGCCAACGATTTGTAGCTAGAAAACCCTTAAAACAACTTATTTAGGGTGGGGATTGGGTATTGGGATTCAACCAAATAAATAATATCGATTGCATAAACTATTAAAAAAAAAAAAACAAGCATTTGAACGATGACTAATTTTTAAAATAACAAATTTTCTATTTATAACTTTATATACCTACTGAATTTTTACGTACATATAACATATTAAAAATGAATAATTTTATAGCACGCCCATCAATAATACCAACAAACGCAGCTGCATATAAACGGCCTCAG **************************************************....((((...))))...((((((.(((((((.(((((((........)))))))((((((((((..((((((......))............((((.(((..........................))).)))).))))..))))))))))..........))))))).))))))..((((((...((((((....))).))).))))))..........(((((....)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M047 female body |
SRR060670 9_testes_total |
SRR060659 Argentina_testes_total |
SRR060664 9_males_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060681 Argx9_testes_total |
SRR1106717 embryo_6-8h |
V053 head |
SRR060687 9_0-2h_embryos_total |
SRR060674 9x140_ovaries_total |
V047 embryo |
SRR060685 9xArg_0-2h_embryos_total |
M027 male body |
M028 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................AGCCAACGATCTGTAGCT..................................................................................................................................................................................................................................................................... | 18 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............AGCGTAGCGCACCATCGGGC............................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................AGCCAACGATCTGTAGCTGG................................................................................................................................................................................................................................................................... | 20 | 2 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGTTGAGCCAACGATTTGTAGCTAGAAAACCCTTTAAACAAC.................................................................................................................................................................................................................................................. | 45 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................AGTTGAGCCAACGATTTGTAGT...................................................................................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................ATCGGGCTATGAATACACCA.................................................................................................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TTAGGGTGGGGATTGGGTATTGGGA..................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GGTGGGGATTGGGTATTGA....................................................................................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................ACCAACAAACGCAGCTGCATAT............ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........GCCCAAGCGTAGCGCACCATCGG................................................................................................................................................................................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................ATACACCAAACGTAAGTTGAGC.................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................AATACGCCAAGCGTAAGTTGA...................................................................................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGCCAACGATCTGTAGCT..................................................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGCCAACGATCTGTAGC...................................................................................................................................................................................................................................................................... | 18 | 2 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................AGCCGACGATCTGTAGCTGG................................................................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.18 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AAGCCGACGATCTGTAGCT..................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ..............................................................................................................................................................................................................................................................................................................TAGCTACAAACGCAGCTGC................ | 19 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................ATTTATGGTGGGGAGTCGG............................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................................................AAAAAAAAAGAAAACAAGCATTT.......................................................................................................................................................... | 23 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....GCACGTCCAAGGGTAGCGC......................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CATTAACTCTTAAAAAAAAAAA..................................................................................................................................................................... | 22 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AGTAGCGTACGGGTTCGCATCGCGTGGTAGCCCGATACTTATGTGGTTTGCATTCAACTCGGTTGCTAAACATCGATCTTTTGGGAATTTTGTTGAATAAATCCCACCCCTAACCCATAACCCTAAGTTGGTTTATTTATTATAGCTAACGTATTTGATAATTTTTTTTTTTTTGTTCGTAAACTTGCTACTGATTAAAAATTTTATTGTTTAAAAGATAAATATTGAAATATATGGATGACTTAAAAATGCATGTATATTGTATAATTTTTACTTATTAAAATATCGTGCGGGTAGTTATTATGGTTGTTTGCGTCGACGTATATTTGCCGGAGTC
**************************************************....((((...))))...((((((.(((((((.(((((((........)))))))((((((((((..((((((......))............((((.(((..........................))).)))).))))..))))))))))..........))))))).))))))..((((((...((((((....))).))).))))))..........(((((....)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060658 140_ovaries_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060681 Argx9_testes_total |
M047 female body |
V047 embryo |
SRR060687 9_0-2h_embryos_total |
GSM1528803 follicle cells |
M061 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................................................GTGCGGGTAGTTATTATGGTT............................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TACGGGTTCGCGTCGCGTGGT..................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................TAAACATCGATCTTTTGGGAA.......................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................TATGGATGACTTAAAAATGCA.................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................ATTATGGTGGTTTGGGTC.................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................CCGGGTAGTTCTTATAGTTGT........................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................ATATCGTGCAGCCAGTTAT.................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................TGTTCAGACACTTGCTACTG................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................GGGTAGTTCTTATAGTTGTGT......................... | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................CCAGCCCTAACCCATTA......................................................................................................................................................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................GAGCCAGATCCTTATGTG.................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................TCGGGTGGTAGCGCGATG............................................................................................................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................................................................................................................ATTATGGTGGTTTGGGTCA................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:19340831-19341167 - | dvi_7957 | TCA------TCGCATGCC---CAAGCGTAGCGCACCATCGGGCTATGAATACACCAAACGTAAGTTGAG---CCA-ACGA------------TTTGTAGCTAG------------------------------------------------------AAAACC------CT------------TAAAA-----CA-----------------------------------------------------------A------CTTATTTAGG--GTGGG----GAT-----TGGGTATTGGGATT--------------------C-------------------------------------------AACCA-----AATAAATAATATCGA---------------------------------------------------------TTGCAT-AAAC---------T-ATTAAAAAA--------AAAAAAACAA-GCATTTGAA--------CGATGACTAATTTTTA--A-------AAT---AACAAAT------TTTCTATTT-A-TAACT--------TTA--TATACCTACTGAATTTT-TACGTACATA---TAACATATTA-AAAAT------GAA-------TA-ATTTTATAGCACGCCCATCAATAATACCAACAAACGCAGCTGCATATAAACGGCCTCAG |
| droMoj3 | scaffold_6496:13039058-13039447 - | TCA------TCGCATGGC---CAAGCGTGGCGCACCATCTGGATATGAATACACCAAACGTAAGTTAAG---CCA-ACGA------------TTCGTAGCGAGTA------------TACCTTAGAAATCCCAACTATGAAC--------------------------------------------AA-----CAACATAACAATGGGGGATCTTCTCT--TAGTGCGGGTCGATACGAAATCTAA-----ATCG------T--TATAAATCAAATTG----ATTGCATTATCTA--TG--------------CAA--------AT-----GTC-----TA----------------------------------------------------------------------------------------------------------------------------------------------------AATATTAAAATGCAGTTGAGCTAACTGATGATGACTAATTCTTATGC-------AAT---AACAAACTAT--CTCTCTACATA--------TAATAAC-TC--TATACCTATTCAATTTA-TATAAATGTA---TAACATATTT-AAAAT------GCA--T----AACATTTTATAGCACGCCCATCAATAATACCAACAAGCGCAGCTGCATATAAACGGCCACAG | |
| droGri2 | scaffold_15245:6841568-6841949 - | TCA------TCGGATGCC---CAAGCGTGGAGCACCATCAGGATATGAATACAACAAACGTAAGTGGAT---------G----CATTTTA--TTTGTGGT-TGTG------------TTTCTTTCAGATCCTTG-TATAAGTCAATACTTATTTTTGGAGGGT------AT----------------------TTC---------------------CTGTTTTT---------TTTTTTACCT--------TGA------T-AATAAAGTAATATTG----ATTGCATTTTGGA--C--------------------------------------------------------------------------------------AAACAGTG--------------------------------------------------------------------CA-A----TTG-T-TATAACA----------TATAATAATGCATTTGAG-------GCGATGACTAATTTTT-----------ATAAATAACAAAA------TTTCTATTT-ATGAACTA-TATAAC-TA--TATACCTATTCAAATTG-TTGATAACTA---TAACATATTA-AA------AATGCA--T----AA-ATTTTATAGCACGCCCATCAATAATACCAACAAACGCAGCCGCATATAAACGACCTCAG | |
| droWil2 | scf2_1100000004510:62292-62662 + | GCA------TCGTATGCCTATAAAACGAGAGCCACACGCCGGATATCAGTACAGCAAACGTAAGTTCTT-TGACA-C-------------------------------------------------------------------------AGTTTCCGTATTT------AG------------TTCATTCTATCAA---------------------------------------CA-CTACAAACA---ACGTT------C---CCCACCCATCCCG----GTAGCATCTTGATAATGGGTAT-CTA---AGTAT--------AT-----ATT-----TAAA------------------------TATAA--------------TAAAATATTCAAAACTTAACCAAATTAAACTTATCTATATATAACAAACAAAAAAAAGAAAAGAAA-TACCAAA-A------AAT-ATTA-------------------------CAAAAAAT-------GAAAAGAAAAAA----------------AT------------------------------------------AA--TAAACCTTAACTAAATA-TAAACATATA---TAATATA-------------------------TA-ATTTTATAGCACGTCCATCAATAATACCAACCAATGCGGCTGCTTATAAACGTCCACAA | |
| dp5 | 3:14834636-14835024 + | GCATCCACATCGTATGCCTCCCAAGCGTGGCCCACCCACCGGCTATGAGTACAGCAAACGTAAGTGGAGTTGATA-ACG----CCTTT------------------------------TG----------TTTA-CAGTCGTAG-TTCGTATAGACTAGATCT------AT------------TAGA-----TCGC---------------------C-----------------TCTTTTCC--------CTCA------C--TCTAAATTCTAGGG----GTCGCACTTTCGA--TGGGTTTCGTT---GGGGT--------AG-----TTT-----GTAG------------------------C-----------------ACAAGTGATTGA-AACT---------------------------------------------------------GAA-A----CTT-C-TAAAGCAAATAAAA----------------AAGA-----------ATGACTAATTTTA---T-------AAT---AACAATCAAA--CGTTTTATATT--------TAACG--ACC--TAAACCATT-----TTT-GAAACACGCA---TAACATACTA-AAAGTAATAATGAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAATGCAGCGGCTTATAAACGTCCACAG | |
| droPer2 | scaffold_4:4217142-4217533 - | GCATCCACATCGTATGCCTCCCAAGCGTGGCCCACCCACCGGCTATGAGTACAGCAAACGTAAGTGGAGTTGATA-ACG----CCTTT------------------------------TG----------TTTA-CAGTCGTAG-TTCGTATAGACTAGATCT------AT------------TAGA-----TCTC---------------------------------------CT---ATTTTCC----CTCA------C--TCTAAATTCTAGGG----GTCGCACTTTCGA--TGGGTTTCGTT---GGGGT--------AG-----TTT-----GTAG------------------------CAC--------------AACAAGTGATTGA-AACT---------------------------------------------------------GAA-A----CTT-C-TAAAGCAAATAAAA----------------AAGA-----------ATGACTAATTTTA---T-------AAT---AACAATCAAA--CGTTTTATATT--------TAACG--ACC--TAAACCATT-----TTT-GAAACACGCA---TAACATACTA-AAAGTAATAATGAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAATGCAGCGGCTTATAAACGTCCACAG | |
| droAna3 | scaffold_13266:7457202-7457632 + | dan_1865 | GCA------CCGTTTGCCCCCAAAACGC---CCGCCCACGGGCTACGAGTATAGTAAACGTAAGTGGT----CCC-TTGCACGCATTCTTT-----------CCG------------ATC----------TTTT-CATCCTTAGAGAATTAATGCTGGGATCC------GT------------TAGATTGGATCCG--------------------------------------CCTGCAGCCTCCA---GTTCA------CCTTTTACATTTTGCGG----GTTGCACTTTTGA--TGGGCTTCCTTTTGGGGGT--------AG-----ACAGTATTGTAG-----------------------------------------AACCAGTGAATGA-AACA----------------------------------AAA---ACAAAAGAAAAAACCAAA-A------AAT-CTTCATGCA---------ATAATATAATACTTTGGA-----------ATGAATAATTTTA-----------AAT---AATAACCA-----TTTGTATATT--------TAATG--ATC--TAAACCATT-----CTTAATTATACACA---TAACCAGTTT-AAAGTAATAATGAAAATCAATGA-ATTTTATAGCACGTCCATCAATAATACCAACAAGCGCTGCGGCTTATAAACGT------ |
| droBip1 | scf7180000396384:270712-271133 + | GCA------CCGTTTGCCCCCAAAACGC---CCGCCCACGGGCTACGAGTATAGTAAACGTAAGTGGT----CCC-TTGCACGCATTCTTT-----------CCG------------ATC----------CTTT-CATCCTTAGAGAATTAATGCTGGGATCC------GT------------TAGATTGGATTCG--------------------------------------CTTGCAGCCTCCA---ATTCA------CCTTTTAGATATTGCGG----GTTGCACTTTTGA--TGGGCTTCCTT-TGGGGGT--------AG-----ACA-----GTAG-----------------------------------------AAACAGTGAATGA-AACA----------------------------------AAA---ACAAA------AAC-AAA-AAATAAGT-TGCAATATGCA---------ATAATATAATACTCTGGA-----------ATGACTAATTTTA-----------AAT---AATAACCA-----TTTGTATATT--------TAATG--ATC--TAAACCATT-----CCTAATTATACGAA---AAACCTCTTA-AAAGTAATAATGAA--TCAATGA-ATTTTATAGCACGTCCATCAATAATACCAACAAGCGCTGCGGCTTATAAACGT------ | |
| droKik1 | scf7180000302476:1750401-1750824 + | GCA------CCGTTTGCCCCCCAAGCGC---GCACCCACGGGCTATGAGTACAGCAAACGTAAGTGGCA-TGACC-AAGCATGATTT---T-----------TTGCCCTCTAGCTACATT----------TTTT-CAGCCTTAGAT---------------CTACATATATAATTATATCTATTAGA-----TCCC---------------------------------------TTCTTTCCATCCTCATTGCATTCACTCCATT--TA-TTTATGGGGC-ACAGCACTTTGAA--TGGGTTTCCTT---GGGAT--------AG-----ATT-----GTAG------------------------CAACA---------CAGAACCAGTGA---------------------------------------------------------ATGAAC-GAGAA-AT--GCTT-CTCAATGCAA--------ATAACATAATTTACTAGA-----------AAAATT----TTC-----------AA----AATAAACTAA--AAT---ATCTT-TATTC--TAATG--ATCGATAAACTTTT-----TT---AAAT---AA---TAACTGACTA-AAAATAATAATGAA--C----AA-ATTTTATAGCACGTCCATCAATAATATCAGCAAACGCAGCGGCATATAAACGA------ | |
| droFic1 | scf7180000454066:2214840-2215247 - | GCA------CCGTATGCCCCCCAAGCGT---CCACCTACGGGCTATGAGTACAGCAAACGTAAGTTGGGTCGCTC-ATGCATGCTCT---T-----------CTA------------TCC----------CTCT-CAGCTGTAGATTCGTAGTGCCGGGGTCT------AG------------TAGTTAAGATCGC---------------------------------------CT-CTGCCTTCC---ACGCA------CCATTCCAATTCTATGG----ATTGCACTTTGGC--TGGGGTACCTC---GGGTT--------AA-----ACT-----GTAG------------------------TATCAAG----TACCAGTAACAGTGA---------------------------------------------------------ATGAAC-AAGAA------CTT-CTCAATGCAAAAAAAAAAAGAATATAATACTCTGGA-----------ATGATTAATTTTA-----------AAT---AACGCAAAAACCTTTTCTATAT-AGTAACG--------------------------------ATACACGCA---TAAACTACTA-AAAATAATAATGAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGT------ | |
| droEle1 | scf7180000490713:154053-154459 + | GCA------CCGTATGCCCCCCAAGCGT---GCACCCACGGGCTATGAGTACAGCAAACGTAAGTGGAG---CCC-TTGCATGCCTT---T-----------CTA------------ATT----------CTTT-CAGCCGTAGATTCGTAGAGCCGGGGTCT------AT------------TATATAAAATCGA---------------------------------------CTCTTACCTTCC---ACGCA------CTCCCCAAGTCTTGTGG----GTTGCACTTTGGT--TGGGATTCCTT---GGGCA--------AG-----CTT-----GTAG----AG------------------TATCAT-----TACCAATACCAATAA---------------------------------------------------------ATGAAC-AAGAA------CTT-ATTAATGCAA-------AAGTATGTAATACTTTGTA-----------ATGATTTATTTAA-----------AAT---AACAAACAAC--TTTTTTATAT-AGTAATGA--------TT--------------------GATACACGCAACGTAACTTACTA-AAAATAATAATGAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCAACAAACGCAGCGGCATATAAACGTCC---- | |
| droRho1 | scf7180000780011:13523-13918 - | ACA------CCGTATGCCCCCCAAGCGT---CCACCCACGGGCTATGAGTACAGCAAACGTAAGTGGAG---CCC-ATGCATGATTT---T-----------CTA------------ATT----------CTTT-CAGCCGTAGATTCGTAGTGCCGGGGTCT------AC------------TAGATAAGATTTC---------------------------------------CCTTTGCCTTCC---ACGCA------CTCCCCAATATTTTTGA----GTTGCACTTTGGA--TGGGGCTCCTT---GGGTC--------AT-----ATT-----GGAG------------------------TATCAG-----TACCAATACCAGTGA---------------------------------------------------------ATGAAC-AAGAA------CTT-CTCAATGCAA-------AAATATATAATACTCTTGA-----------ATGATTAATTTTA-----------AAT---AACGAAATAC--TTTTTTATAT-AGTAATA--------------------------------ATACACGCA---TAACTTACTA-AAAATAATAATGAA--A----CA-ATTTTATAGCACGTCCATCAATAATACCAACAAACGCAGCGGCATATAAACGT------ | |
| droBia1 | scf7180000302143:2402535-2402940 + | GCA------CCGTATGCCCCCCAAGCGT---CCACCCACGGGCTATGAGTACAGCAAACGTAAGTGGTG---CAA-ATGCATGCTAT---T-----------CTA------------TCC----------CTTT-CAGCCGTAGATTCGTAGAACCGGGGTCT------AT------------TAGATAAAAGCGC---------------------------------------TTTCTACCTTCC---G-------------TCCCCGTTTTGTGGACCGTTTGCACCTTTGA--TGGGGCTCCTT---GGGTT--------AT-----ATT-----GTAGTAGCAG------------------AACCAG-----TACCAATACCAATAA--------------------------------------------------TCA----ATGAAC-AATAA------CTT-CTCAATGCAA-------AACTATATAATACTCTGGA-----------ATGATTAAATTTA-----------CAT---AACGAACAAC--TTTTTTATAT-AGTAATGA------------------------------TATACACGCA---TAACATACTA-AAAGTAATAATGAA--C----AA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGTCC---- | |
| droTak1 | scf7180000415739:163525-163947 - | GCA------CCGTATGCCCCCCAAGCGT---CCACCCACGGGCTATGAGTACAGCAAACGTAAGTGGAG---CCC-ATGCATGTTTT---T-----------CTG------------TTC----------CTTT-CAGCCGTAGATTCGTAGTGCCGGGGTCT------AT------------TAGATAAGATCGC---------------------------------------TTTCCGCCTTCC---AAGCA------CTGTCCCAATTAAAGGG----GTTGCACTTTGGA--TGGGGCTCCTT---GGGTT--------AG-----ATT-----GTAGTATCAGTATCAGTACCTGTTCCGATACCAG-----TACCAAAACCAGTGA---------------------------------------------------------ATGAAC-AATAA------CTT-CTCAATGCAA--------AATATATAATACTCTGGA-----------ATGATTAATTTTA-----------AAT---AACAAAAAAC--TTTTTTATAT-TGTAATGA------------------------------TATATGCGCA---TAACATACTA-AAAATAATAATAAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGTCC---- | |
| droEug1 | scf7180000409672:6211128-6211500 - | GCA------CCGTATGCCTCCCAAGCGC---CCACCCACAGGCTATGAGTACAGCAAACGTAAGTTGAG---ACCAATGCATACTTA---A-----------CTG------------CTC----------CTTT-CAGCCGTAG--------TGTCGATA--T------ATTACACAATC-ATTAGATAAGATTGC---------------------------------------TTA--------C---ACAAA------CAATCCCAGTTTTATGG----GATGCACTTTGGA--TGGGGTTCCTT---GGGTT--------AG-----ATT-----GTAG------------------------TATCAA-----TACCAATACAAGTGA---------------------------------------------------------ATGAAC-AAAAA------ATT-CTCAATGCAA-------AACTGCATAATATTCT-------------------TAATTTTA-----------AAT---AACAAAAAAA--GTCT-T------------A------------------------------TATACTCGCA---TAACATAATAAAAAATAATAATGAA--A----CA-ATTTTATAGCACGTCCATCAATAATATCTACAAACGGAGCGGCATATAAACGT------ | |
| dm3 | chr2R:18297482-18297865 - | ACA------TCGTATGCCCCCCAAGCGT---CCGCCTACGGGCTATGAGTACAGCAAACGTAAGTTAAT---CCC-AAGCATGCATTCTTT---------------------------------------------------ATTTGGGTAGTGCCGGGGTCT------AT------------GACATAAGATCGA---------------------------------------TTTTTCCCCTGA---ATGCA-----------CCAGTTTTTTGG----GATGCACCTTTGA--CAGGA-ACTTT---GGGTTATAGATT-AGATTAGATC-----ATAC------------------------TATCAA-----AACCAATAACTGTGA---------------------------------------------------------TTGAGC-AAGC---------T-CTCAGATCA--------AACTATATAATACACTGGA-----------ATGATTAATATTA-----------TGC---AACGATTCAC--TCCTT--TAT-AGTAATTA------------------------------CATACACGCA---TAAAATACTA-CAAATAATAATCAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGT------ | |
| droSim2 | 2l:20501645-20501975 - | dsi_17093 | ------------TATGCCCCCCAGTCAT---CCGCCTACGAGCTATGAGTACAGGAAACGTAAGTAT------------------------------------------------------------------------------TTGGTAGTGCCGGGGTCC------AT------------TACATAAGATTG---------------------------------------------------------------------------TTTTATGG----GATG--------------------------GGATATAGATTTAGATACGAT-------TAT------------------------TATCGAGTATTAACTGATAACAGTGA---------------------------------------------------------ATGAGC-AAGA---------T-CTCAGATCA--------AACTATATAAAACACTAGA-----------ATGACTAATATTA---TAATATTATAC---AACGATTAAC--TTCTT--TAT-AGTAATTA------------------------------CATACACGCA---TAAATTACTA-CTAATAATAATCAA--C----CA-ATTTTATAGCACTTCCATTAATAATACCGGAAAACGCAGTGGCATATAAACGTCCATAT |
| droSec2 | scaffold_9:1605529-1605894 - | ACA------TCGTATGCCCCCCAAGCGT---CCTCCTACTGGCTATGAGTACAGCAAACGTAAGTTGAT---CCC-AAGCATGCATTCTTT---------------------------------------------------ATTTGGGTAGTGCCGGGGTCT------AT-----A------AACATAATATAGA---------------------------------------TTTC-CCCCTAA---AGGCA------CTTCGCCAGTTTTTGGG----GAT---------------------TT---GGGTTATAGATT-AG-----ATC-----ATAC------------------------TATCAA-----AACCAATAACAGTGA---------------------------------------------------------TTGAGC-AAGC---------T-CTCAGATCA--------AACTATATAATACACTGGA-----------ATGATTAATATTA-----------TGC---AACGATTCAC--TTCTT--TAT-AGTAATTA------------------------------CATACACGCA---TAACTTACTA-CAAATAATAATCAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCTGCGGCATATAAACGT------ | |
| droYak3 | 2R:18047221-18047580 - | ACA------TCGTATGCCTCCCAAGCGT---CCCACTACGGGCTATGAGTACAGCAAACGTAAGTGGAA---CCC-AAGCATGCATTCTTAAT---------TT---------------------------------------GTTTCTTAGTGCCGGGGCCT------AT------------TACA--------------------------------------------------------------------------CTCCCCCATTTTGAGGG----GATGCACCTTTGA--TTTGAATC-TT---GGGTCACAGAAT-AG-----ATT-----ATAG------------------------TATCAA-----AACCGATAACAGTGA---------------------------------------------------------ATGAGC-AAGA---------T-CTTGAAACA--------AACTATATAACACTCTGGA-----------ATGATTAATATTA-----------AAG---AACAAACCAC--TTCC---TAT-AGTAATTA------------------------------CATACACGCA---TAAACTACTA-CAAATAATAATCAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGT------ | |
| droEre2 | scaffold_4845:12437916-12438256 - | ACA------TCGTATGCCCCCCAAGCGT---CCTACCACGGGCTATGAGTACAGCAAACGTAAGTGGAA---CCC-AAGCATGCATTCTTA---------------------------------------TTTT-CT--------TTCTTAGTGGCGGGGTCT------AT------------TCC--------------------------------------------------------------------A------CTCCGCCAGTTGTATGG----GATGAACCTTTGA--TTAGAATC-TT---GGGTGT---------------C--------AG------------------------TATCAA-----AACAAATAACAATGA---------------------------------------------------------ATGAGC-AAGA---------T-CTCGAATCA--------AACTATATAAT-CTCTGGA-----------ATGATTAATA--------------------ATCGAACCAC--TTCC---TAT-AGTAATTA------------------------------CACACACGCA---TAAACTACTA-CAAATAATAATAAA--C----CA-ATTTTATAGCACGTCCATCAATAATACCGACAAACGCAGCGGCATATAAACGT------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:44 AM