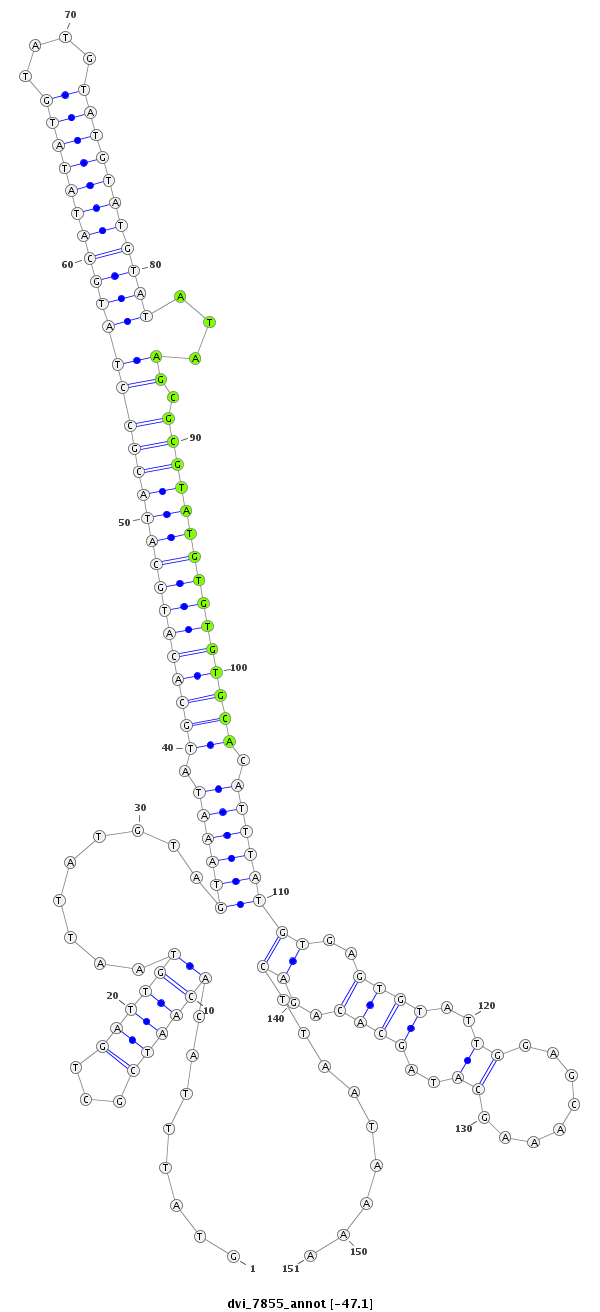
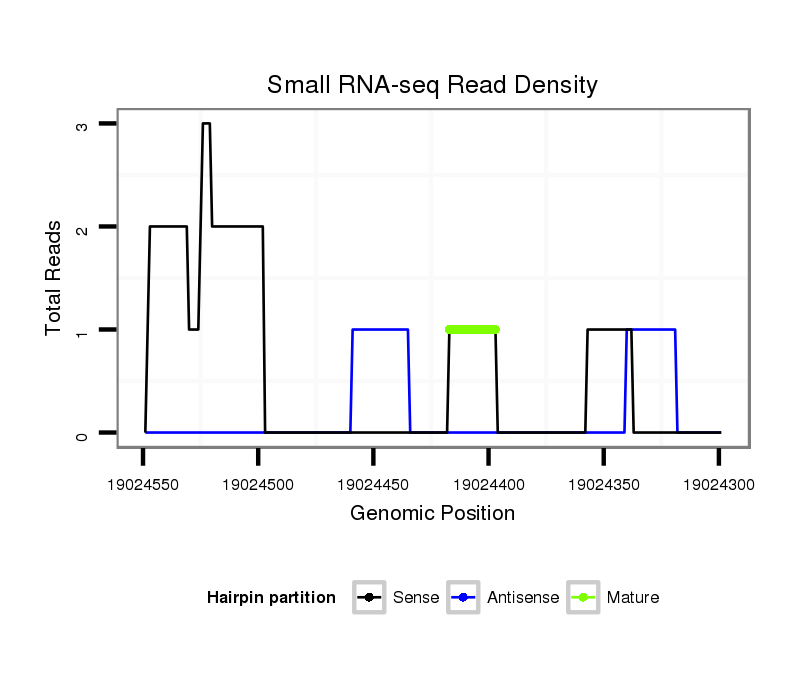
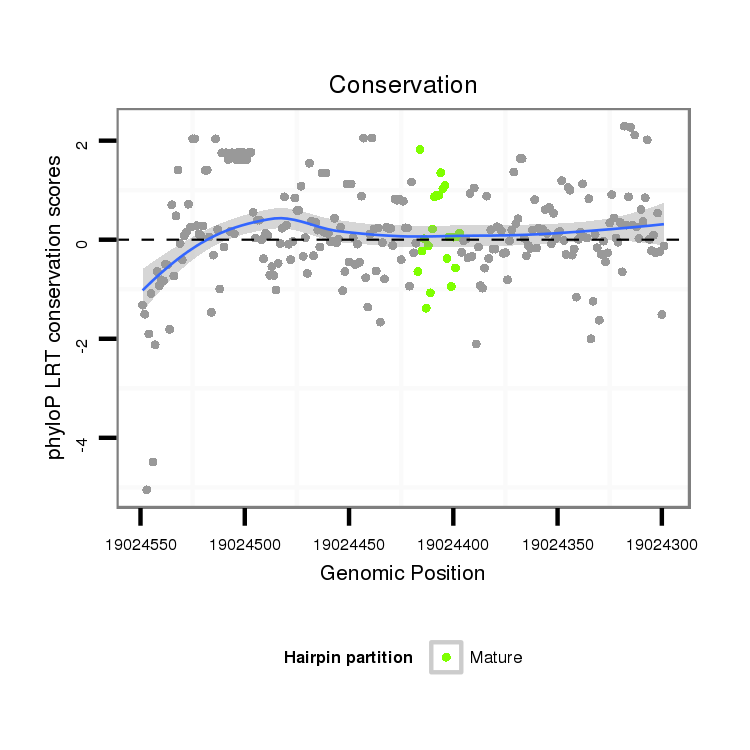
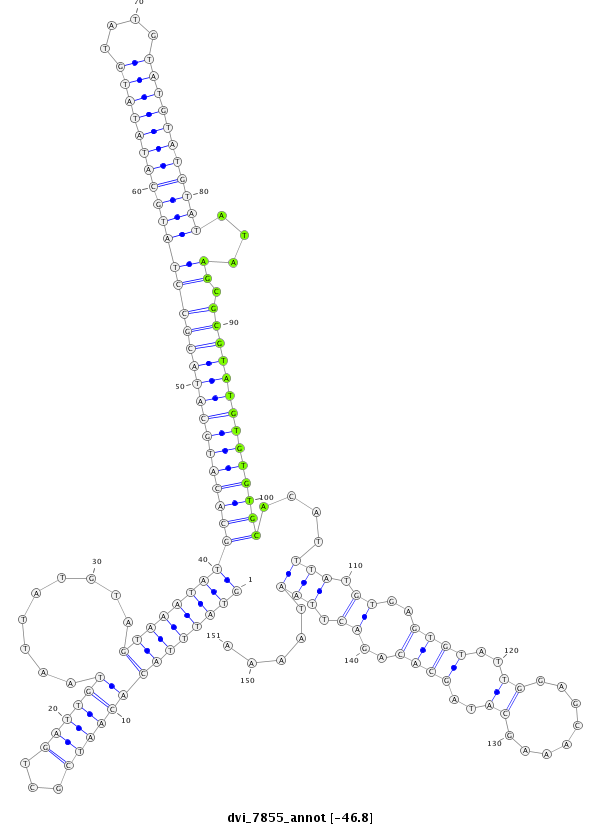
ID:dvi_7855 |
Coordinate:scaffold_12875:19024349-19024499 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -46.8 | -46.7 | -46.6 |
 |
 |
 |
CDS [Dvir\GJ19969-cds]; exon [dvir_GLEANR_5434:1]; intron [Dvir\GJ19969-in]
| Name | Class | Family | Strand |
| (CATA)n | Simple_repeat | Simple_repeat | + |
| Homo6 | DNA | hAT-Pegasus | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCCGCTCCGGAGTCGGACGCAGCAAACCTTTAAAAAAACACCACAGTGAGGTATTTACACAATCGCTGATTGTAATTATGTAGTAAATATGCACATGCATACGCCTATGCATATATGTATGTATGTATGTATATAAGCGCGTATGTGTGTGCACATTTATGTGAGTGTATTGGAGCAAAGCATAGCACAGACTTAATAAAAAACAGCTACAACCGAGCGCCAAGCGACAGTTTCAGTCTGTGCCGTGTGCT **************************************************........((((((...)))))).........((((((.((((((((((((((((((((((((((((....)))))))))))...)).))))))))))))))).))))))((..((((..((........))..))))..)).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
V116 male body |
M027 male body |
M028 head |
SRR060667 160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
M061 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................ACCTTTAAAAAAACACCACAGTGAGGT....................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGCTCCGGAGTCGGACGCAGCAAACCT.............................................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CCGCTCCGGAGTCGGACG........................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................ATAAGCGCGTATGTGTGTGCA.................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................TTAATAAAAAACAGCTACAA....................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................TTGGAGCAAAGCATGGCACA.............................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AACCTTTAAAAAAACACCACAGTGAGGT....................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................ATGTAAGTGTATTGGAGG........................................................................... | 18 | 2 | 14 | 0.21 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................GCAGCCATCCTTTAAAGAA...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................AATGTAAGTGTATTGGAGGAA......................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CAGTTTCAGTGTGTGCCGG..... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CGGTGAGGTATTTGCACAGT............................................................................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................ACACAATCGCTGTTTATGAT............................................................................................................................................................................... | 20 | 3 | 18 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| .....................................................................................................................TAAGTATGTATGTATATAAGTGC............................................................................................................... | 23 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................GCAGCCATCCTTTAAAGAAA..................................................................................................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CGGCGAGGCCTCAGCCTGCGTCGTTTGGAAATTTTTTTGTGGTGTCACTCCATAAATGTGTTAGCGACTAACATTAATACATCATTTATACGTGTACGTATGCGGATACGTATATACATACATACATACATATATTCGCGCATACACACACGTGTAAATACACTCACATAACCTCGTTTCGTATCGTGTCTGAATTATTTTTTGTCGATGTTGGCTCGCGGTTCGCTGTCAAAGTCAGACACGGCACACGA
**************************************************........((((((...)))))).........((((((.((((((((((((((((((((((((((((....)))))))))))...)).))))))))))))))).))))))((..((((..((........))..))))..)).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
M047 female body |
SRR060666 160_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................GTTGGCTCGCGGTTCGCTGTCA.................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................CGTGTACGTATGCGGATACGTATAT........................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................AGTAATTTTTTTGTGGTG............................................................................................................................................................................................................... | 18 | 2 | 20 | 0.30 | 6 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...........................GTAATTTTTTTGTGGTGG.............................................................................................................................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AATTATTCTGTGGTCTCACT.......................................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CGTATATACATACATACATAC.......................................................................................................................... | 21 | 0 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................ATTCGTGTGCGTATGCGG.................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................TACATACATACATACATATAT.................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:19024299-19024549 - | dvi_7855 | GCC-GCTCCGGAG-TC--------------------GGACGCAGC--AAA---------------CCTTTAAAAAAACACCACAGTGAGGTATTTAC---ACAAT-CGCTGATTGTAAT-TATGTAGTAA---A--------------TATGCACAT----GCATACGCC--------------TATGCATATATGTAT----GTATGTATG----------------------------------------------------------------------------------------------------------------------TA----TAT--------------AAGCGCGTATGTGTGTGCACATTTA-------------------------------------------------------------TGTGAGTGT----------------------------------------ATTGGAGCAAAGCATAG-CACAGACTTAATAAAA---------------------AACA-GCTACAACCGAGCGCCAAGCGAC-AGTTTCAGT---------------------------------------------------CTGTGC-CGTGTGCT |
| droMoj3 | scaffold_6496:12707587-12707901 - | GCG-GCTCCGGTG-TC--------------------ATACGCAGC--GAA---------------TCTGTACGAAAGCGCCACAGTGAGGTATTTAC---TCAAT-CTCTGATTGTAAT-TATGTAGTAA---ATAATATTTACTAAATATGCACAA----GCACACA--AAAACACACAGCCATACACAAACCCAACT----GTATACATATGTGTCTATTGTGTATGCCCGAGTGTTT----------------------------------------------------------------------------G--TGTC-------------------------------TC------TGTGTG----TGTGTG----------------TGTGC-------CTGTGCATATTTAATAAATAG--AA----TTTATGTAAGTG---------------------------------------CTGCGGGCAAAAAGCATAG-CCCAGACTTAATAAAG-------C-----------------------A----GCCGCCCAGCGAC-AGTTTCAGT---------------------------------------------------CTGTGC-CGTGTGCT | |
| droGri2 | scaffold_15245:5086032-5086310 - | GCA-ACCGCGATC-GC--------------------A---GCAGC--GAA---------------TCTTTACGAAAACACCACAGTGAGGTATGATGATCACAAT-CGCTGATTGTAAT-TATGTAGTAA---A--------------TATGCACAT----ACATACGCACATACATACATACATACATACATACATAC----ATACATACACA--------------------------------------------------------------------------------------------------------------------------AATTCATACATACATCC--------------ACATAGACGTACATATATATGTACATATGT---------ATTTGCATATT------------TA--------TTTGCTTGT----------------------------------------GCTGGAA--AAGCATTG-CACAGACTTAATAAAA-A--------------------------------------CCTGGCGACAAGCTTCAGT---------------------------------------------------CTGTGC-CGTGTGCT | |
| droWil2 | scf2_1100000004513:944193-944402 - | GCG-GCAGCAGCA-GC--------------------A---GCATT--AAAAAACAAACAAA---ATCGTTACGAAAACGCCACAGTGAGGTAGGATGATGATTGGATGATAATGAAA-------------A------------------------------------------------------------------------------------TGATTGGATT---------------------------CTGATTGTATTCTGATTGGCCTATGTATTTGACCCACTACCAACTGTGTAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACAGACTAAATATAAAGCCCGAC-----------------------A----TCGGAGCAATGTT-AAGTGCAGT---------------------------------------------------TTGTGC-CGTGTGCT | |
| dp5 | 3:3675699-3675938 + | TCT-ATTCGTGAG-CC-----------------------CGTATC--AAA-TCGA----------------ACCACCCGCCACAGTGAGGTATTATC---GCTAA-TTCGGATTGTAAT-TGGCTGCT----------------------------------------------------------------------------------------------------------------------------------------------------GCACT-CACTCACTACCCCGTGTGCATCCCATAACTATATG--TGT------------------------------------------------------------------------------------CCTATCT----------------AA--------TCTGGGTGC-------------------------------------TCTGCTGGAAACGAGCATAGACCCAGTCT------GAGGTCCAGTTTCAGTCATCAATAAAA-GGCACTAAC-GGCTCCCATCGAT--GCTGCAGT---------------------------------------------------CTGTGC-CGTGTGCT | |
| droPer2 | scaffold_2:3862053-3862292 + | TCT-ATTCGTGAG-CC-----------------------CGTATC--AAA-TCGA----------------ACCACCCGCCACAGTGAGGTATTATC---GCTAA-TTCGGATTGTAAT-TGGCTGCT----------------------------------------------------------------------------------------------------------------------------------------------------GCACT-CACTCACTACCCCGTGTGCATCCCATAACTATGTG--TGT------------------------------------------------------------------------------------CCTATCT----------------AA--------TCTGGGTGC-------------------------------------TCTGCTGGAAACGAGCATAGACCCAGTCT------GAGGTCCAGTTTCAGTCGCCAATAAAA-GGCACTACC-TGCTCCCATCGAT--GCTGCAGT---------------------------------------------------CTGTGC-CGTGTGCT | |
| droAna3 | scaffold_13266:5002989-5003129 + | TAA-ATCTATATAATATACTCCTCCTATACATATGTA---TGC------------------------------------------------------------------------------------------------------------------------------C--------------TACATAT--------------GT-------------------------------------------------------------------------------------------------------------C--TGTC-------------------------------TC------T--------------------------------------------------TCTT------------TA--------TGTACGTGTGTTAGTTGGAGG---CCATGGAATATAAGGCAAGCAA-------------------------------------------------------------------------TCCGTCCGGCGAT--GCTGCAGT---------------------------------------------------CTGTGC-CGTGTTCT | |
| droBip1 | scf7180000395396:16898-16992 + | TAA-TTATTAATT-AA--------------------------------------------------------------------------------------------------------TAT----------------------------GTATA--------------------TATGTATATATGTATATATGTAT----ATATGTATATA--------------------------------------------------------------------------------------------------------------------------TGTATATATGTATATAT--------------ATGTATATGTA-------------------------------------------------------------TATGTGTAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT | |
| droKik1 | scf7180000302385:906579-906811 - | CCG-AAA--------T-----------------------CGTATC--AAA-TCAA----------------ATCATCCGCCGCCGTGAGGTATTATC---GCCCC-AACTGATCATCAT-CGGT------GTG---------------------------------------------------------------------------------TTCCCAGTGTTCGTGTTCCAGTCTCCGTGTCCCCCCGAGTGTCCATA-----------------------------------------ATCCATAATCCAGTA--TGT--------------T--------------GTGTGTGT--------------GTGTG--------------------TTTGAGGTGTCTGAGTG--------------------------G---------------AGGCCACCAT-GAATATAAAACATGCAA----------------------------------------------------------------TC--------CCTGCCGGCGAT--GCTGCAGTTA-------------------------------------------------CAGCGC-CGGGTGCT | |
| droFic1 | scf7180000453784:823292-823399 + | GGA-GCTTAGCAT-TC--------------------A---GAATC--AAA-TACTTAAATACCCTTGGTTAAATATT-------------------C---GCTAT-TGTGGAATATAAT-TGTATACAAG---A--------------AACACCTAT----TTAAAATAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTCAAAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT | |
| droEle1 | scf7180000491232:173989-174221 - | CCG-AGA--------T-----------------------CGTATC--AAA-TCAA----------------ATCATCCGCCGCCGTGAGGTATTATC---ATTGCCCAGTGATCGGA-------------G------------------------------------------------------------------------------------TCCCAGTGTT---------------------------GTGTCCATAATCC-ATAACCATGTGTACATGTTCCGGTGGCGGCAGTATGGGT-----------CTGTGTTCCTGGAGTGTG--T--------------GTGTGTGT--------------GTGTG---------------------------------------------------TG--------TGAACGTGTGT-----GCCGG---CCAT-GAATATAAAACATGCAA-------------------------------------------------------------------------TCCGTCCGGCGAT--GCTGCAGT---------------------------------------------------CTGTGCCGGGTTACT | |
| droRho1 | scf7180000776638:16510-16632 - | TAT-GTATATGTA-TA--------------------------------------------------------------------------------------------------------TGT------------------------------------------------ATATGTATATGTATATGTATATGTATAT----GTATATGTATA--------------------------------------------------------------------------------------------------------------------------TGTATATGTATATGTAT--------------ATGTATATGTATATGTATATGTATATGTAT---------AT--------------------------------------------------------------------------------------------TATAA-TATATATATAA--------------------------------------------------------------------------------------------------------------------------------TA | |
| droBia1 | scf7180000301232:32878-32978 - | TGT-GTATATGTA-TG--------------------------------------------------------------------------------------------------------TGT------------------------------------------------ATATGTATATGTATATGTATGTGTATAT----GTATGTGTATA--------------------------------------------------------------------------------------------------------------------------TGTATATGTATGTGTAT--------------GTGTATATGTATATGTATATGTATAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CG | |
| droTak1 | scf7180000415313:154613-154740 - | ATA-CACA-TAGA-TA-------------------TA---TAC------------------------------------------------------------------------------------------------------------------------------C--------------TATATATATTTGAAT----GTATGTATG----------------------------------------------------------------------------------------------------------------------TG----GTT--------------AAACA----------------------------------------------ACAAGTAAATG-------AA---GTGATCTA----T-----TAC----------------------------------------ATTATTGTAAAATATAT-CAAAAAACTGAGAAAA---------------------A------------------AA--------------------------------------------------------------------CAACAC-AAACAAAC | |
| droEug1 | scf7180000408984:903637-903805 + | TGT-ACATATGTA-TG--------------------------------------------------------------------------------------------------------TAA------------------------------------------------ATATGTATGTACATACGTATGTACATTC----ATATGTATGTA--------------------------------------------------------------------------------------------------------------------------CATACGTATGTACATTC--------------ATATGTATGTACATACGTATGTACATACAT---------ACTTACATACA------------TA--------TTTAC-------------------------------------------------------ATATTT-CATATACATTGAACAA-A--------------------------------------CACTAAAACCATTTTCAT------------------------------------------------------------TTCCTTT | |
| dm3 | chr2R:15560488-15560667 - | ATA-TTTA-TATA-TA-------------------TA---TAT--------------------------------------------------------------------------------------------------------------------------------A------------TATATATATATATAT----GTATGTATG----------------------------------------------------------------------------------------------------------------------TA----TAT--------------ATATGTGTATGTATGTATATATATG-------------------------------------------------------------TAT-----------------------AT-GTATGTATAAAATGCAA----------------------------------------------------------------TTCGTACTGATCGGTAAGCGTT-TGT---AG---AGTGCAGCCGCGTGCTCTTGGCGAAGCGATTGCATCTTTTGGCTTTGTG------CAGTTTACT | |
| droSim2 | x:11538751-11538844 + | ATT-TTAA-AA-------------------------------------------------------------------------------------------------------------TAT----------A--------------TATACACT----TGT--------A------------TATGCATGCGTATATGTATGCATATATG----------------------------------------------------------------------------------------------------------------------TA----TGT--------------ATGTGTGTATGTGCAAGCACACTCA-------------------------------------------------------------GGAAACTGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT | |
| droSec2 | scaffold_1616:3863-3996 + | ATC-TGCGCA---------------------------------------------------------------------------------------------AT-GTATGTCTCTAATATGTGTAATGT---A--------------TA----------TGG--------A------------GATGTATGCGTATAT----GTATGTATA---------------TGGCGATG-----------------------------------------------------------------------------------------------TATGCGT--------------ATATGTATGTA----------TATGTA----------------TGT---------ATTTATATG--------------------------G--------------------------------------------------------------------------------------------------------------------------------A--GATGTAAT---------------------------------------------------GCATGC-AATGTATA | |
| droYak3 | 2R:17246172-17246310 - | AAACATTCGAAAA-AC-----------------------CAAAGCAAAAA-TTAA----------------ACAATACAC-ACA-------------------CT-GTACTACTCCACACTAT----------------------------GTATGTATCTAC--------A------------TACATATATATGTAT----GTATGTATG----------------------------------------------------------------------------------------------------------------------TA----TGT--------------ATGTATGTATGTATGTATATATGTA-------------------------------------------------------------TGTGATTAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT | |
| droEre2 | scaffold_4770:12436134-12436320 - | TGT-ATGTATGTA-TG--------------------------------------------------------------------------------------------------------TAT------------------------------------------------GTATGTATGTATGTATGTATGTATGTAT----GTATGTACGTA--------------------------------------------------------------------------------------------------------------------------TGTATGTATGTATGTAT--------------GTATGTACATATGTATGTATGTATGT-------------ATGTACATATG------------TG--------T----------------------------------------------------------------G-TTTGGTCATAATCAGC---------------------ATCAGGTTGCTACCGAAAACCAAACGAT-CGTTG------GT------------------------------CACCTTCTGGAATT----------CCTGTGCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 11:23 PM