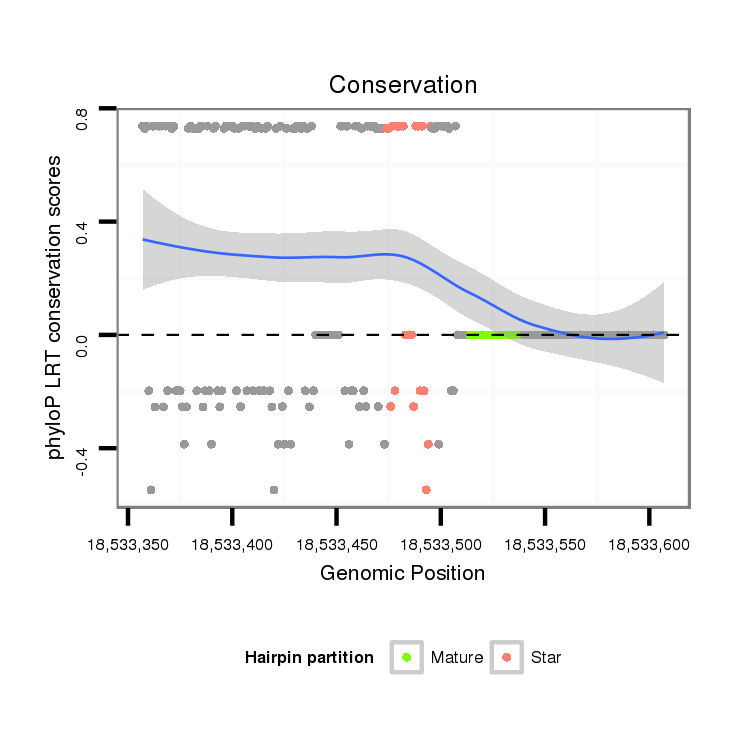
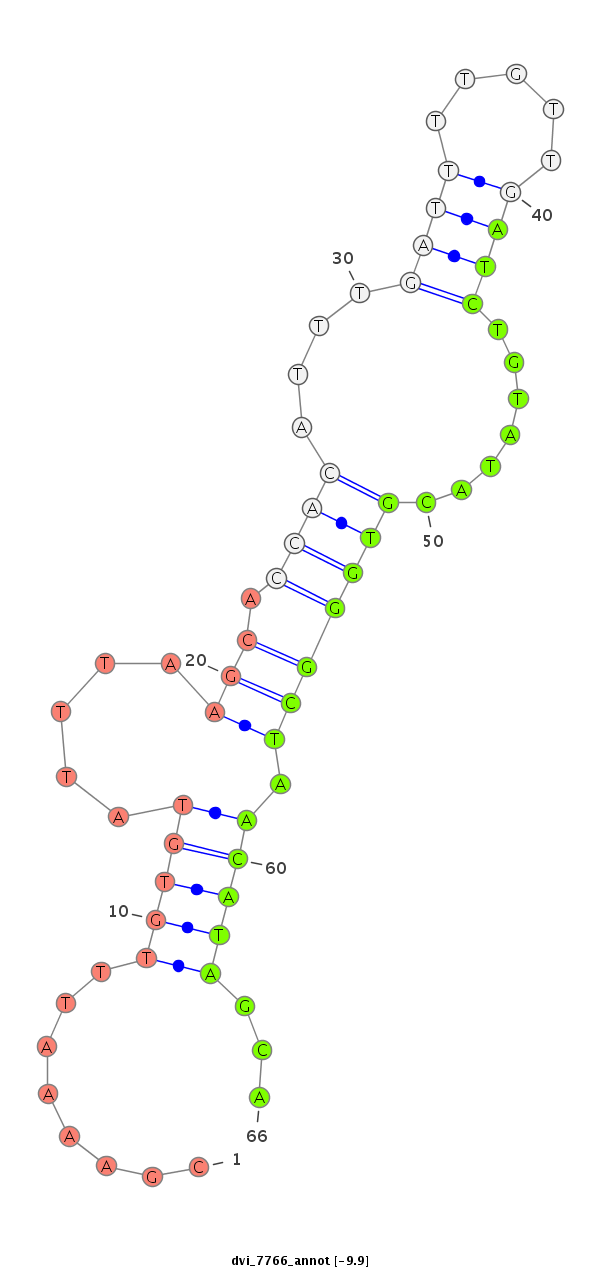
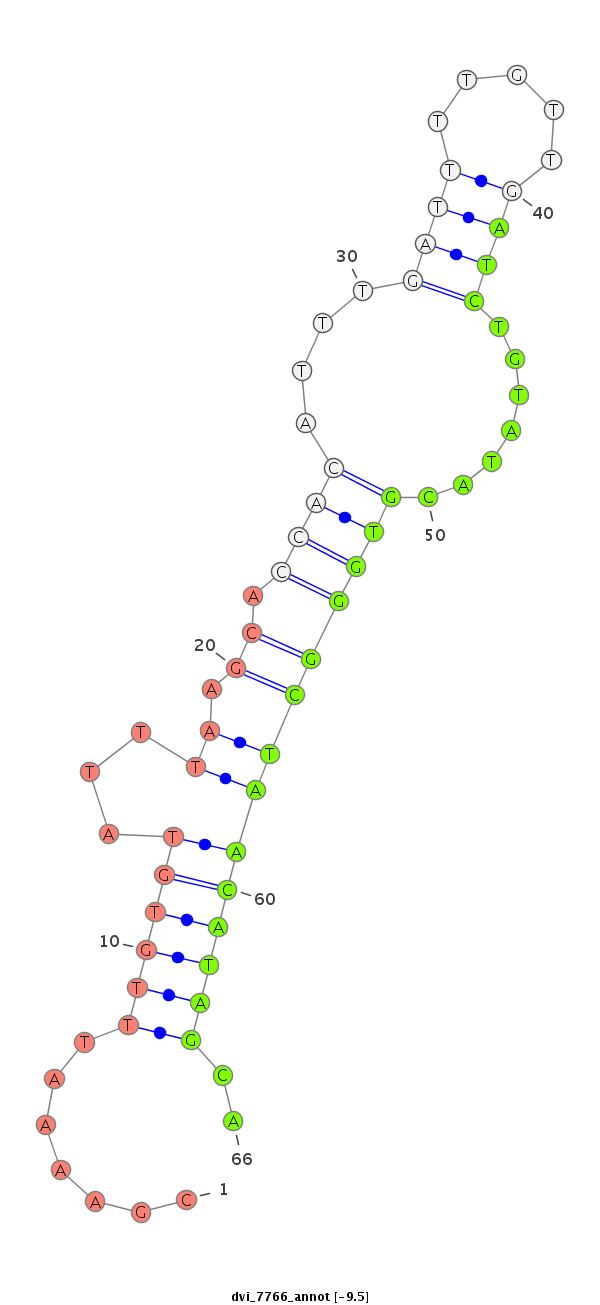
ID:dvi_7766 |
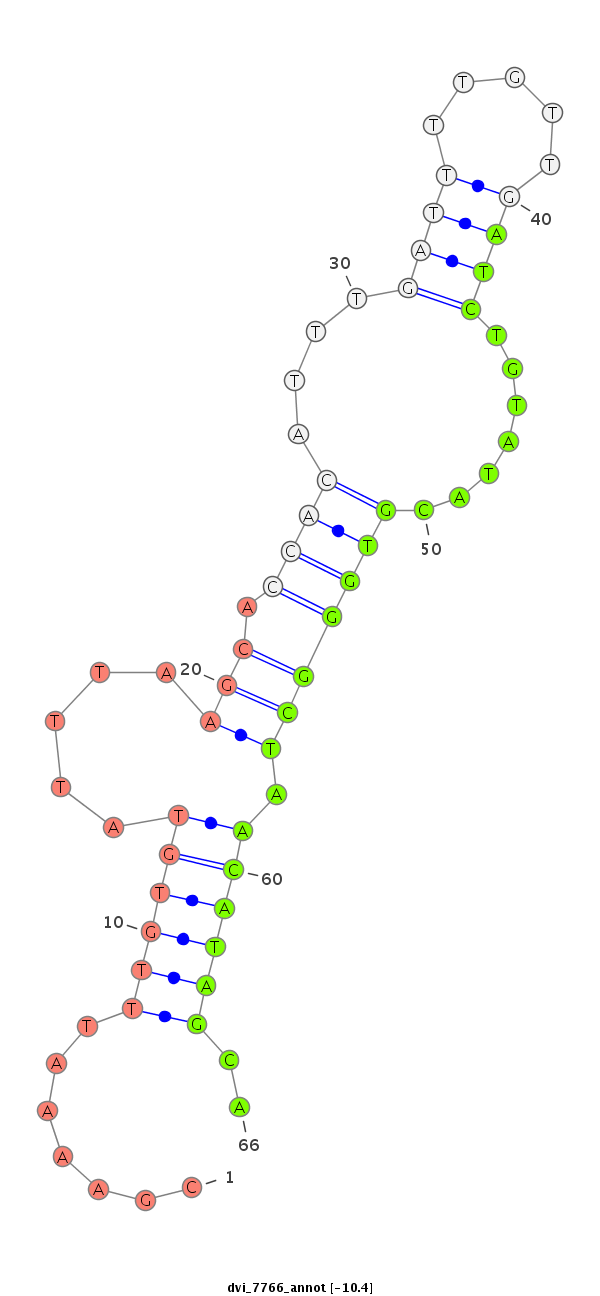
Coordinate:scaffold_12875:18533407-18533557 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
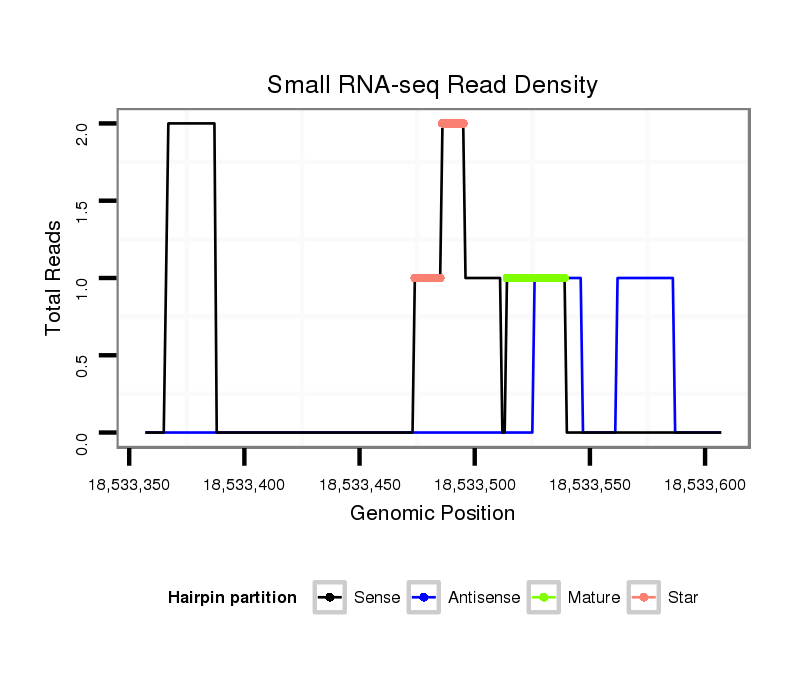
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.9 | -9.5 |
 |
 |
CDS [Dvir\GJ22357-cds]; exon [dvir_GLEANR_7649:1]; intron [Dvir\GJ22357-in]
No Repeatable elements found
| ------------------------------------------########--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGAAGATAATTAAAGAGACTGTCAGCGGATATACGAATCGAAATGGGATTGTTAGTAGGCATTGCCAAAGTAGGTACTAGTAACGCGTAATATTATATACGTAAAGAAAATACAGCCCGAAAATTTGTGTATTTAAGCACCACATTTGATTTTGTTGATCTGTATACGTGGGCTAACATAGCATCGTTTTAATTTCAATACCTCAAAGGATATTCTCTGCCAATTGTAATAATATTTGAGAAAGAGAAAGA *********************************************************************************************************************.......((((((.....(((.((((....((((.....)))).......))))))).))))))..******************************************************************** |
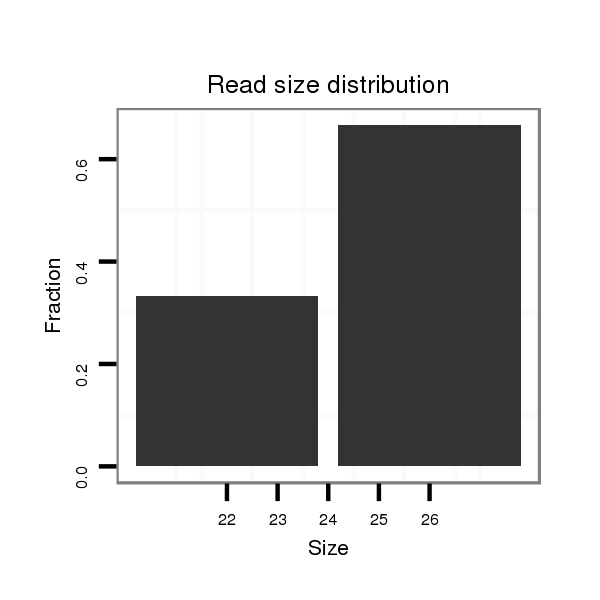
Read size | # Mismatch | Hit Count | Total Norm | Total | M028 head |
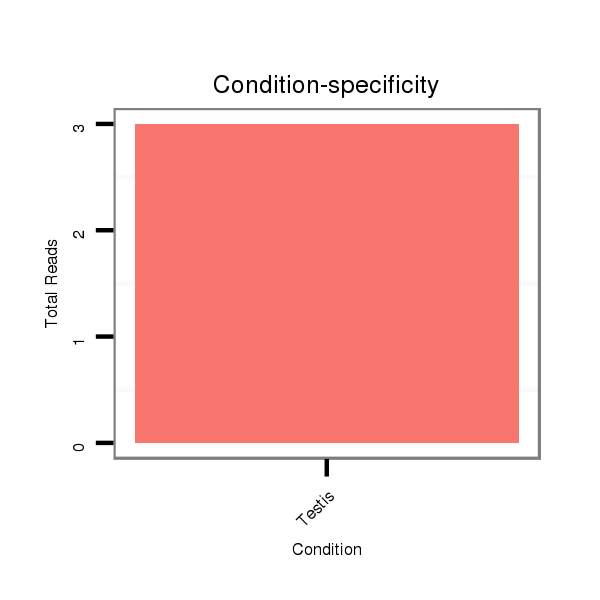
SRR060678 9x140_testes_total |
V053 head |
M047 female body |
SRR060659 Argentina_testes_total |
SRR060667 160_females_carcasses_total |
SRR060680 9xArg_testes_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........TAAAGAGACTGTCAGCGGATA............................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TTAAAGAGACTGTCAGCGGATA............................................................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................ATCGAAATGGGATTGTTGGTA.................................................................................................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................CGTGGGCTGACATGGCATCGT................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................ATCTGTATACGTGGGCTAACATAGCA.................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CGAAAATTTGTGTATTTAAGCA................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TATTTAAGCACCACATTTGATTTTGT................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......AATTAAGGAGACTGTCAGCGG............................................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................TGGGATTGTTGGTAGGCATTG........................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TAATAATGTTAGGGAAAGAGA.... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................GGATAGGTAGTAGCCATTG........................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACTTCTATTAATTTCTCTGACAGTCGCCTATATGCTTAGCTTTACCCTAACAATCATCCGTAACGGTTTCATCCATGATCATTGCGCATTATAATATATGCATTTCTTTTATGTCGGGCTTTTAAACACATAAATTCGTGGTGTAAACTAAAACAACTAGACATATGCACCCGATTGTATCGTAGCAAAATTAAAGTTATGGAGTTTCCTATAAGAGACGGTTAACATTATTATAAACTCTTTCTCTTTCT
********************************************************************.......((((((.....(((.((((....((((.....)))).......))))))).))))))..********************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060674 9x140_ovaries_total |
M047 female body |
SRR060676 9xArg_ovaries_total |
SRR060678 9x140_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR1106714 embryo_2-4h |
SRR060675 140x9_ovaries_total |
V047 embryo |
M027 male body |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................TAAAGTTTTGGAGTTTCATAT....................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AAAGTTTTGGAGTTTCATAT....................................... | 20 | 2 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................CCCGATTGTATCGTAGCAAAA............................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AAAGTTTTGGAGTTTCATATAAG.................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TTCCTATAAGAGACGGTTAACATTA..................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TTAAAGTTTTGGAGTTTCATATGAGA................................... | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AAAGTTTTGGAGTTTCATA........................................ | 19 | 2 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TAAAGTTTTGGAGTTTCATATGA..................................... | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................TAAAGTTTTGGAGTTTCATA........................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AAAGTTTTGGAGTTTCATATG...................................... | 21 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AAGTTTTGGAGTTTCATAT....................................... | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................TCGTATAAGAGACGGT............................. | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TTAAAGTTTTGGAGTTTCA.......................................... | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................ATAAATTCGTACAGTAAACTAA.................................................................................................... | 22 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................CTTGACCCCAACAATCCT.................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................AAAATTATGGAGTTTGCT......................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:18533357-18533607 + | dvi_7766 | TGAAGATAATTAAAGAGACTGTCAGCGGATA-TACGAATCGAAATGGGATTGTTAGTAGGCATTGCCAAAGTAGGTACTAGTAACGCGTAATATTATATACGTAAAGAAAATACAGCCCGAAAATTTGTGTATTTAAGCACCACATTTGATTTTGTTGATCTGTATACGTGGGCTAACATAGCATCGTTTTAATTTCAATACCTCAAAGGATATTCTCTGCCAATTGTAATAATATTTGAGAAAGAGAAAGA |
| droBip1 | scf7180000396249:668-803 - | TGAGCAAAATAAGAGAAGTATACAGCAGAAGCTGAGAGATGAAATGAGTTTATTGGTGGACACACCAATCGCCGGTACTGGAAG------------TACAAACAATGGTAATACTGCACGTAGATTT----TTTCAGCAACCAAATTTGGCT---------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/16/2015 at 09:28 PM