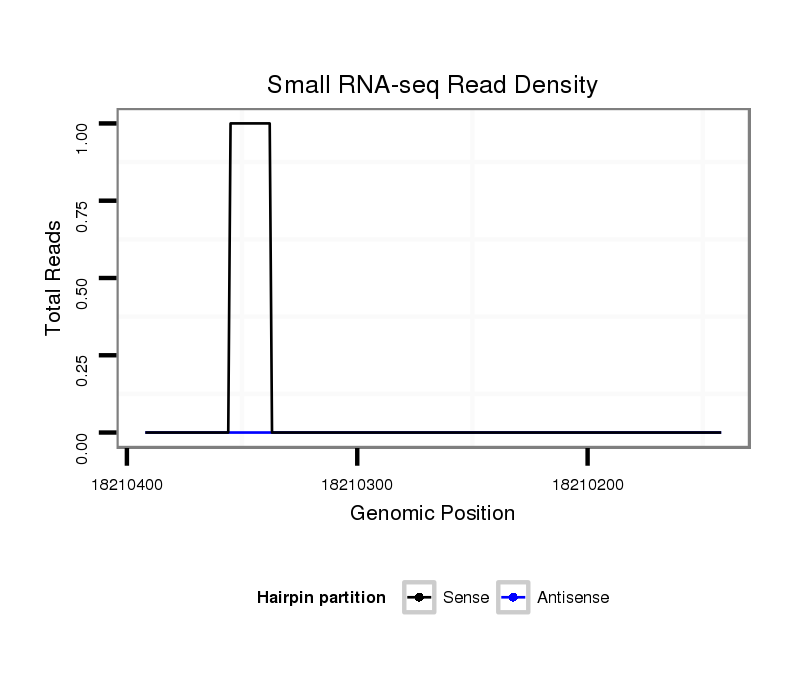
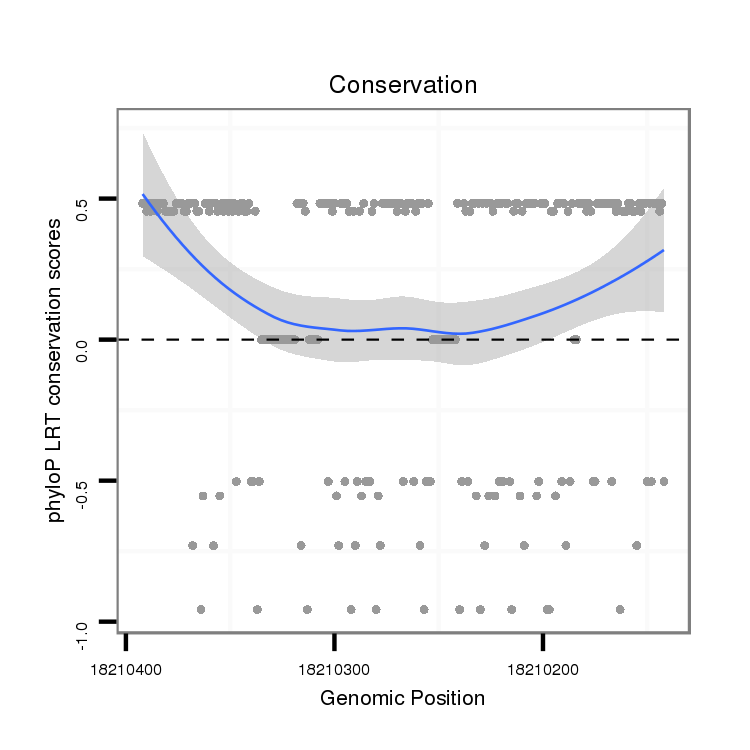
ID:dvi_7688 |
Coordinate:scaffold_12875:18210192-18210342 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ20017-cds]; exon [dvir_GLEANR_5479:1]; intron [Dvir\GJ20017-in]
No Repeatable elements found
| -----#############################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATGAGATGTTACGCGCTATTCGAACTCGGTTTGAATCATGTCAGAACAAGGTGTGCTTTTTAATTGAATACAATTACAGCTATTTATAACTCATAAACACGCCCCTTGGCGACATATTAATTGTTAGTTATCTCTGAATTTAAGTATTGTATCAAGCGATAACACAATCAAGTAAAAGCAATGCAATTAATATTCGGCAAACTTGTTTTTTATCAGTGATTAAATGTGACGCGAAGCGTGTTCTCAATCAA **************************************************(((((((...((((((....)))))).)))................)))).....(((.(((.(((((((((((..(((....((..(((....((((((.....))))))....)))..))....)))...)))))))))))))).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
V116 male body |
SRR060681 Argx9_testes_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ................................TAATCCTGTCAGTACAAGGTGT..................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................ATGTCAGAACAAGGTGTG.................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................CGAAGCATGATCTCATTCAA | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................ATTAAGATTCCGCAACCTT............................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
TACTCTACAATGCGCGATAAGCTTGAGCCAAACTTAGTACAGTCTTGTTCCACACGAAAAATTAACTTATGTTAATGTCGATAAATATTGAGTATTTGTGCGGGGAACCGCTGTATAATTAACAATCAATAGAGACTTAAATTCATAACATAGTTCGCTATTGTGTTAGTTCATTTTCGTTACGTTAATTATAAGCCGTTTGAACAAAAAATAGTCACTAATTTACACTGCGCTTCGCACAAGAGTTAGTT
**************************************************(((((((...((((((....)))))).)))................)))).....(((.(((.(((((((((((..(((....((..(((....((((((.....))))))....)))..))....)))...)))))))))))))).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060660 Argentina_ovaries_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060658 140_ovaries_total |
SRR060672 9x160_females_carcasses_total |
GSM1528803 follicle cells |
M027 male body |
SRR060676 9xArg_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............CGGGATAAGTCTGAGCCAAA........................................................................................................................................................................................................................... | 20 | 3 | 2 | 17.50 | 35 | 9 | 9 | 7 | 2 | 5 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............CGGGATAAGTCTGAGCCAA............................................................................................................................................................................................................................ | 19 | 3 | 9 | 16.44 | 148 | 38 | 28 | 25 | 27 | 15 | 7 | 0 | 1 | 3 | 0 | 3 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................GCCCTTCGAGCAAAAAATAG..................................... | 20 | 3 | 2 | 5.00 | 10 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ...........GCGGGATAAGTCTGAGCCAAA........................................................................................................................................................................................................................... | 21 | 3 | 1 | 4.00 | 4 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GCGGGATAAGTCTGAGCCAA............................................................................................................................................................................................................................ | 20 | 3 | 3 | 2.67 | 8 | 2 | 2 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................GCCCTTCGAACAAAAAATAG..................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGCGGGATAAGCCTGAGCGAAA........................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GGGATAAGTCTGAGCCAAA........................................................................................................................................................................................................................... | 19 | 3 | 6 | 0.67 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GGGATAAGTCTGAGCCAA............................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.50 | 10 | 4 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........GCGGGATAAGTCTGAGCCA............................................................................................................................................................................................................................. | 19 | 3 | 9 | 0.33 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ATAGTTCGCTATTGTGTT.................................................................................... | 18 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGGGATAAGCCTGAGCTAA............................................................................................................................................................................................................................ | 19 | 3 | 18 | 0.17 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................TTGGTACAGTCGTGTTCCG....................................................................................................................................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ............CGTGATAAGTCTGAGCCAAA........................................................................................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................ATAAGCTTGAGCCTAGCCT........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:18210142-18210392 - | dvi_7688 | ATGAGATGTTACGCGCTATTCGAACTCGGTTTGAATCATGTCAGAACAAGGTGTGCTTTTTAATTGAATACAATTACAGCTATTTATAACTCA----TAAACACGCCCCTTGGCGACATATTAATTGTTAGTTATCTCTGAATTTAAGTATTGTATCAAGCGATAACACAATCAAGTAAAAGCAATGCAATTAATATTCGGCAAACTTGTTTTTTATCAGTGATTAAATGTGACGCGAAGCG-TGTTCTCAATCAA---- |
| droMoj3 | scaffold_6496:11844080-11844303 - | ATGAGATGTTACGCGCTATTCGAAATCGCATTGACTCTTGTCAGAGCAAGGTACGGC-----------------TAAAGG-----ATAATTCAATTAACAATACCCATCATAATGAGTGATTAATTGTTGGTTACCTATCGAC------------TGGAGTGATTAGAAATTCTAATGAAGCCAAAGAAATTATCATTGCGCTAATTGGCT--TTATCAGCAATTAAATATGAGGCGAAGCTTTGTTTTTAATCAGTGTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 08:23 PM