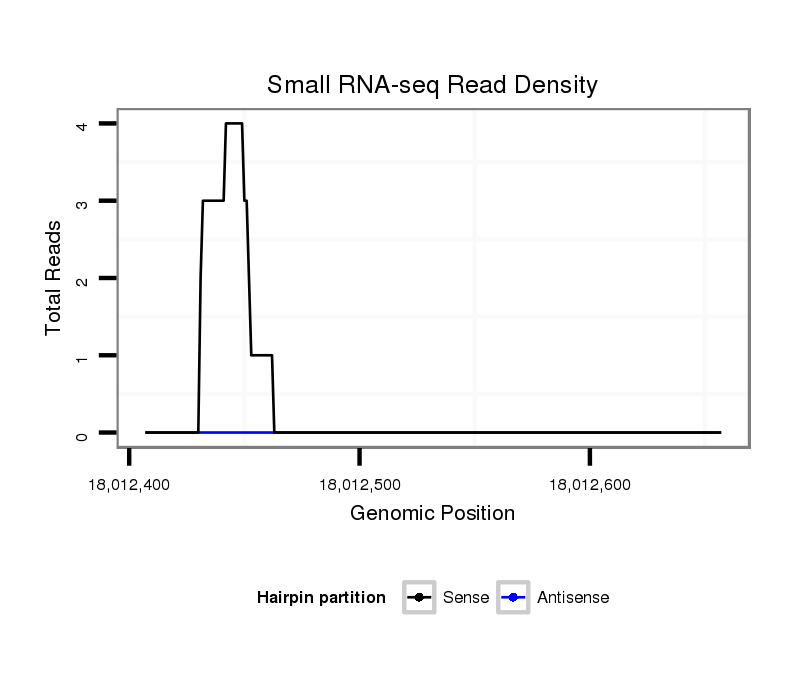
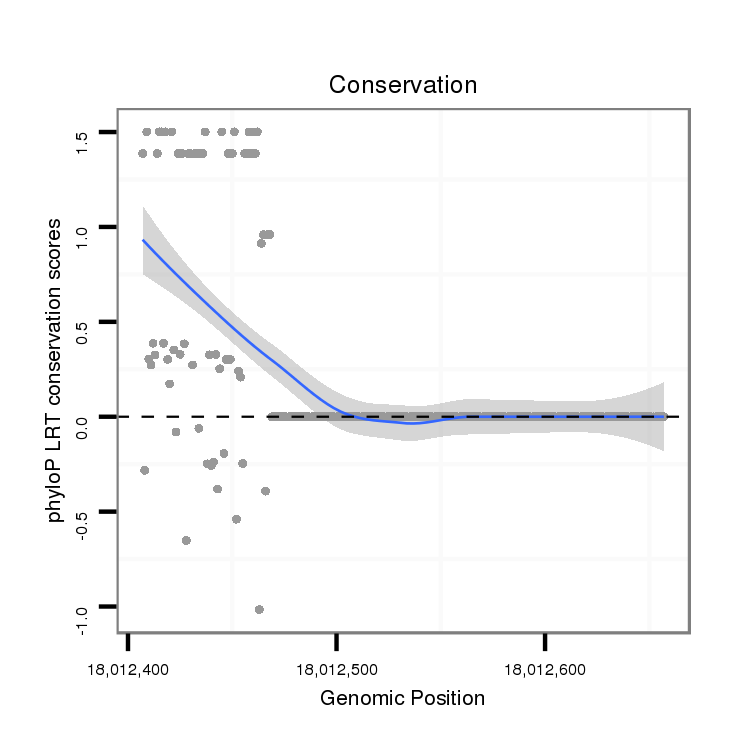
ID:dvi_7625 |
Coordinate:scaffold_12875:18012457-18012607 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ22320-cds]; exon [dvir_GLEANR_7615:1]; intron [Dvir\GJ22320-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CCTGATTCAAATGGAATCTGTGGCAGCTGGACTTCGAGAAGGCCTTTCAGGTGAGTCGAAATGTACAGAATTTATGAAGAAAATCCTGCCTTGAGTGGGCAGACTACCTCAGTTGCTTGAGTTGGTAGATATATACCTGTGTAAATGGCTGCTCTTGGGGGTTTTTTATTCTTAAATAATTGTTTTTCTTGTATGTTAATTTTGAAAAATATTTAACTGACTTAACTGAGTCTGCTAAAAGCCGATAAGTA **************************************************..((........((((((((((((.(((((((((((..((..((((((.((...(((..((((((((......)))))........))).)))..)).))))))..)))))))))))..)).))))).........))).)))).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060660 Argentina_ovaries_total |
SRR060670 9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR1106720 embryo_8-10h |
SRR1106722 embryo_10-12h |
SRR1106728 larvae |
V116 male body |
SRR1106713 embryo_0-2h |
SRR1106716 embryo_4-6h |
V053 head |
SRR060685 9xArg_0-2h_embryos_total |
SRR1106727 larvae |
SRR1106714 embryo_2-4h |
SRR1106718 embryo_6-8h |
SRR1106729 mixed whole adult body |
V047 embryo |
SRR060666 160_males_carcasses_total |
SRR1106723 embryo_12-14h |
SRR1106724 embryo_12-14h |
SRR1106730 embryo_16-30h |
SRR060686 Argx9_0-2h_embryos_total |
SRR060654 160x9_ovaries_total |
M028 head |
SRR1106715 embryo_4-6h |
SRR1106719 embryo_8-10h |
SRR1106725 embryo_14-16h |
SRR1106726 embryo_14-16h |
SRR060673 9_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR1106717 embryo_6-8h |
SRR1106721 embryo_10-12h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................AGTTGCTTGATTTGGT............................................................................................................................. | 16 | 1 | 20 | 4.10 | 82 | 0 | 0 | 0 | 0 | 17 | 11 | 5 | 0 | 8 | 7 | 0 | 0 | 5 | 4 | 2 | 4 | 0 | 0 | 3 | 3 | 3 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 1 |
| ........................AGCTGGACTTCGAGAAGGCCTT............................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCTGGACTTCGAGAAGGCCT.............................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................GAGAAGGCCTTTCAGGTGAGT................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................AGCTGGACTTCGAGAAGGC................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CAGTTGCTTGATTTGGT............................................................................................................................. | 17 | 1 | 10 | 0.40 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GTTTACCTCCGTTGCTTGAG.................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGTTGGTAGAGCGATACCT................................................................................................................. | 19 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................ATTGTTTTTCTTGGATGT....................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTCTGCCTTGATTGGGCAAA.................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GTTGCTTCATTTGGTAGA.......................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACTACCGCAGTTGGTTTAGT................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TTACAGAATCGATGAAGAAAA........................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCTGGAGTTCGAGAACGC................................................................................................................................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ACCTCAAGTGCTTGATTTGGT............................................................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................ACTGACTTAACTAAGGATG................. | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TTACAGAATTGATGAATAAAA........................................................................................................................................................................ | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................AGGGCAGATCTTGGGGGT......................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GGACTAAGTTTACCTTAGACACCGTCGACCTGAAGCTCTTCCGGAAAGTCCACTCAGCTTTACATGTCTTAAATACTTCTTTTAGGACGGAACTCACCCGTCTGATGGAGTCAACGAACTCAACCATCTATATATGGACACATTTACCGACGAGAACCCCCAAAAAATAAGAATTTATTAACAAAAAGAACATACAATTAAAACTTTTTATAAATTGACTGAATTGACTCAGACGATTTTCGGCTATTCAT
**************************************************..((........((((((((((((.(((((((((((..((..((((((.((...(((..((((((((......)))))........))).)))..)).))))))..)))))))))))..)).))))).........))).)))).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
SRR060668 160x9_males_carcasses_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................TCTTTTGGGACAGAACTTAC.......................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................TCAGACGAGTTTCGGCAACT... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| ..................ACACCGACGACCCTAAGCTC..................................................................................................................................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:18012407-18012657 + | dvi_7625 | CCTG---------ATTCAAATGGAATCTGTGGCAGCTGGACTTCGAGAAGGCCTTTCAGGTGAGTCGA----------AATGTACAGAATTTATGAAGAAAATCCTGCCTTGAGTGGGCAGACTACCTCAGTTGCTTGAGTTGGTAGATATATACCTGTGTAAATGGCTGCTCTTGGGGGTTTTTTATTCTTAAATAATTGTTTTTCTTGTATGTTAATTTTGAAAAATATTTAACTGACTTAACTGAGTCTGCTAAAAGCCGATAAGTA |
| droMoj3 | scaffold_6496:11637470-11637525 + | CCTG---------AATCAATTGGAAACTGCAGCAGCCGGATTTCGAGAGGGCCTTTCGGGTGAGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15245:4151881-4151942 + | CTTG---------ATTCAAATGGAATCTGTGGCAGCTGGATTGAACGAAGGCCTCTCAGGTGAGTGGA----------TAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13266:5060235-5060315 - | CCTGGTGGCCCTTCTCCAAATACAAACAGTCGCCGCCGGAGCTGGCAACAGTCTGCATGGTGAGTAGAATTCGAAATCTAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000395751:866374-866429 - | CCTT---------CTCCAAATACAAACAGTCGCCGCCGGAGCTGGCAACAGTCTGCATGGTGAGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409474:688493-688548 + | CGTG---------CTCCAAATACAGACAGTAGCCGCCGGAGCGGGTGACAGTCTATCAGGTGAGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/19/2015 at 02:34 PM