
ID:dvi_7504 |
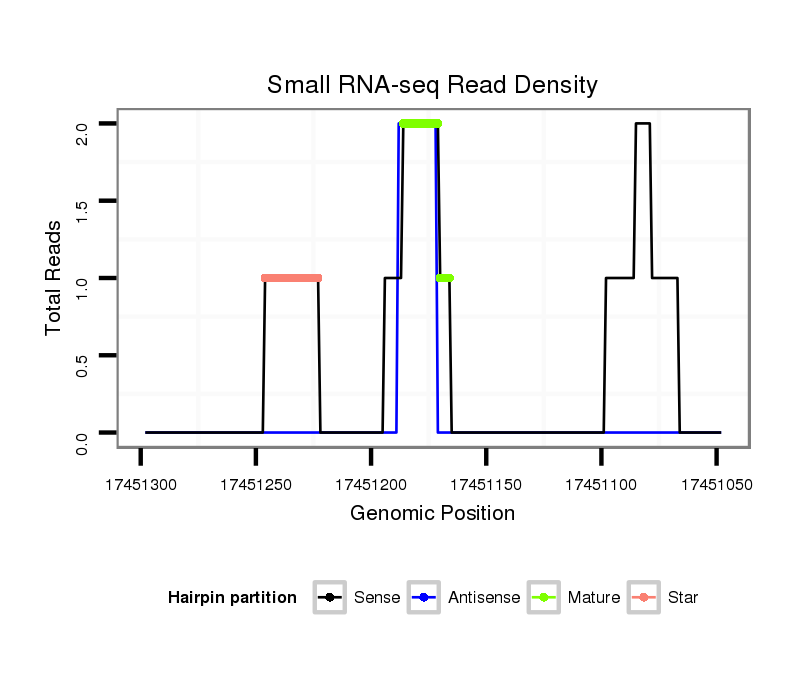
Coordinate:scaffold_12875:17451098-17451248 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -6.7 | -6.0 | -5.7 |
 |
 |
 |
intron [Dvir\GJ20080-in]; CDS [Dvir\GJ20080-cds]; exon [dvir_GLEANR_5538:2]; intron [Dvir\GJ20080-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#######################################----------- TAAACAACAAAAAGAACGCATCGAAAGCAATTGATAAAAAAAGGAGACCCAATAGGACTATCTCTGCTGAAAAGAAACGTGAATCCAATCAGAAAAATGCAACAAAAGCTGCAGACGTTAGAATAAAGGATAAAAGAGGTAGCTAAACTAGTTAAAAAAATATATTTATTTCTTCTGATATTTCTCTCTATATCCTTACAGACGACGAACTCTCGGAGGACTTCGATGAAGAAGCGTATGGTAAGAGTTTT ****************************************************.....(((..(((((..........(((...((......))...)))...........)))))...)))............********************************************************************************************************************** |
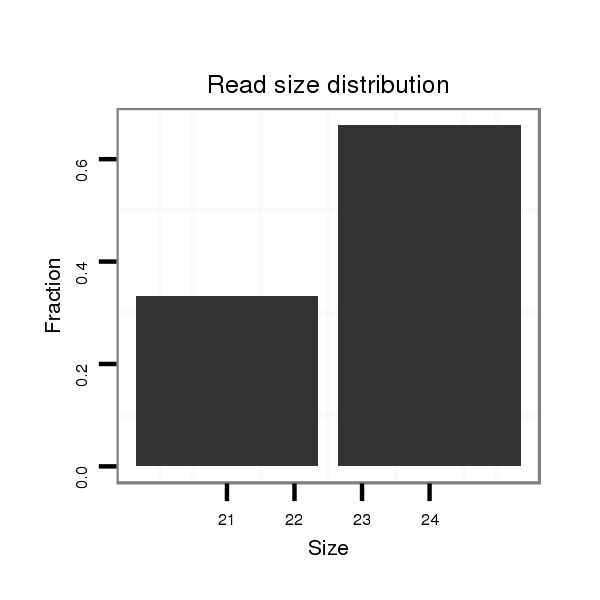
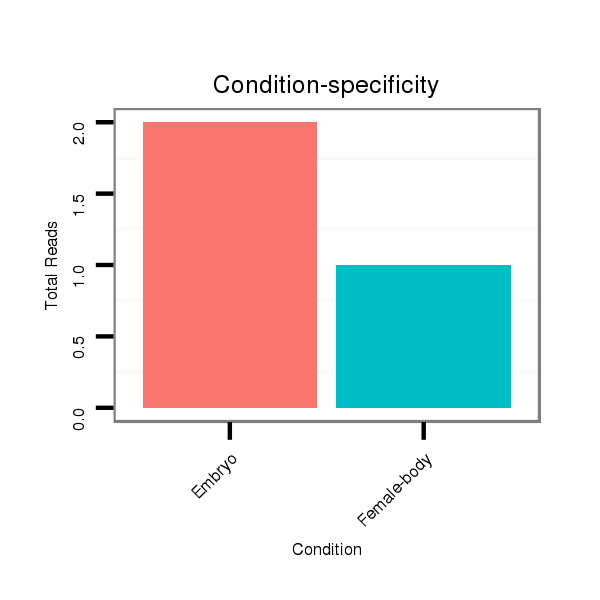
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M047 female body |
V053 head |
M028 head |
SRR060658 140_ovaries_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060670 9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060659 Argentina_testes_total |
SRR060672 9x160_females_carcasses_total |
SRR060689 160x9_testes_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TAAATGATGAAAGAGGTAGC............................................................................................................ | 20 | 2 | 1 | 4.00 | 4 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GACGACGAACTCTCGGAGGA............................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AGACGTTAGAATAAAGGATAA...................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................AAAGCTGCAGACGTTAGAATAAAG........................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CGGAGGACTTCGATGAAGA................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TAGGACTGCGATGAAGAGGCG............... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TTGTTAAAAAGAGGAGACCCA........................................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................GAAGACCCAATAGGACTATC............................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TAGGACTTCGATGAAGAGGCG............... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAGGACTATCTCTGCTGAAAAGAA............................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGTCTGATGAAAAGAAACG............................................................................................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGAAAGAAGCAACAAAACCTGC........................................................................................................................................... | 22 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AATGATGAAAGAGGTAGC............................................................................................................ | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................ATGAAGGAAAAAAGAGGT............................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................GATATTTCTCTTTATTTCC........................................................ | 19 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GTACCATCTCTGCTGAAGA.................................................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................CTTACAAACGACGAACAGT...................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GGGACATCGATGAAGTAGC................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGAATAAAAAATAAAGGAGGT............................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATTTGTTGTTTTTCTTGCGTAGCTTTCGTTAACTATTTTTTTCCTCTGGGTTATCCTGATAGAGACGACTTTTCTTTGCACTTAGGTTAGTCTTTTTACGTTGTTTTCGACGTCTGCAATCTTATTTCCTATTTTCTCCATCGATTTGATCAATTTTTTTATATAAATAAAGAAGACTATAAAGAGAGATATAGGAATGTCTGCTGCTTGAGAGCCTCCTGAAGCTACTTCTTCGCATACCATTCTCAAAA
**********************************************************************************************************************.....(((..(((((..........(((...((......))...)))...........)))))...)))............**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
V116 male body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
M047 female body |
SRR060656 9x160_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060655 9x160_testes_total |
SRR060678 9x140_testes_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................GCTGATTGGGACCCTCCTG.............................. | 19 | 3 | 7 | 6.00 | 42 | 0 | 11 | 9 | 7 | 7 | 4 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................CGTCTGCAATCTTATTT............................................................................................................................ | 17 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................CTTTTCGCTTAGGTCAGTCTTT............................................................................................................................................................ | 22 | 3 | 5 | 0.80 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TCGACGTTTGTAATCTTATTG............................................................................................................................ | 21 | 3 | 11 | 0.27 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| .................................................................................................................................................................................TATAAAGAGAGCTAGAGGACT..................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CTGATTGAGACCCTCCTGC............................. | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CTGATTGGGAGCCTCCTGC............................. | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:17451048-17451298 - | dvi_7504 | TAAACAACAAAAAGAACGCATCGAAAGCAATTGATAAAAAAAGGAGACCCAATAGGACTATCTCTGCTGAAAAGAAACGTGAATCCAATCAGAAAAATGCAACAAAAGCTGCAGACGTTAGAATAAAGGATAAAAGAGGTAGCTAAACTAGTTAAAAAAATATATTTATTTCTTCTGATATTTCTCTCTATATCCTTACAGACGACGAACTCTCGGAGGACTTCGATGAAGAAGCGTATGGTAAGAGTTTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
Generated: 05/17/2015 at 05:12 AM