
ID:dvi_7423 |
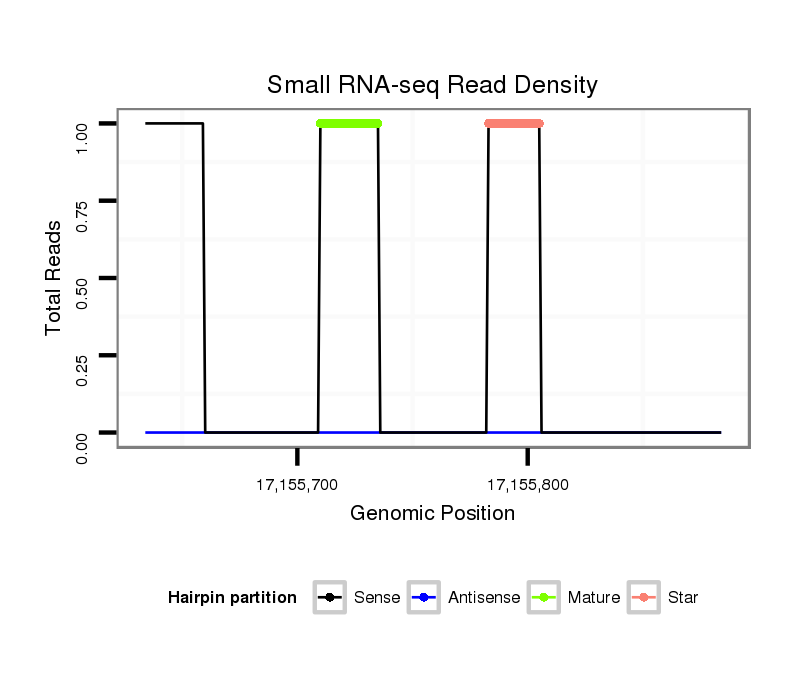
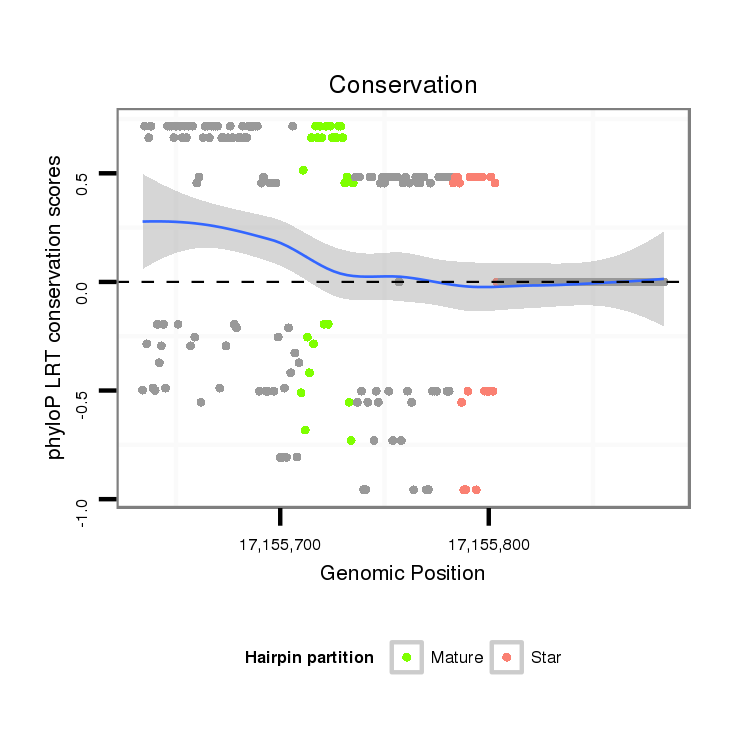
Coordinate:scaffold_12875:17155684-17155834 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -17.8 | -17.6 | -17.6 |
 |
 |
 |
CDS [Dvir\GJ22258-cds]; exon [dvir_GLEANR_7559:6]; intron [Dvir\GJ22258-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TAACTGAGGCGCATTGTGACTGTCTTGTTGTTGATATGCCCGAGGTGGAGGTAAAAACAGCCCACAGGGGTATCCGTTCAACAAGTGCAGTGCGATCGATAGATAACCTAATTTGAGAAACAATTCTCCAGAGCTAGCCATGTTTAGAAGATCTGCGATTGTTTACCATGTAGATCAGTGATCCCTGCGCTGGCATTTTAGCTGTGATGAGAGAAAGGTGCATCATGCGAAAGATAAGATCTGAAAAGAAG ****************************************************************************...(((((.((((....((((..((((((..(((.((((((((....)))).))))..)))...))))))...)))))))).))))).........******************************************************************************* |
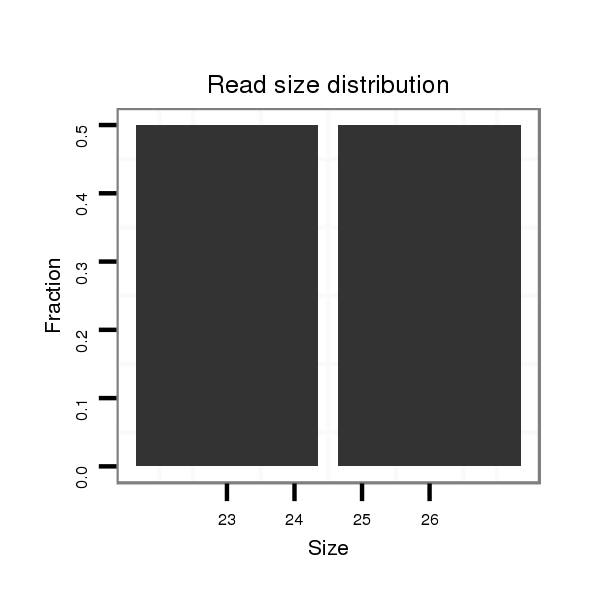
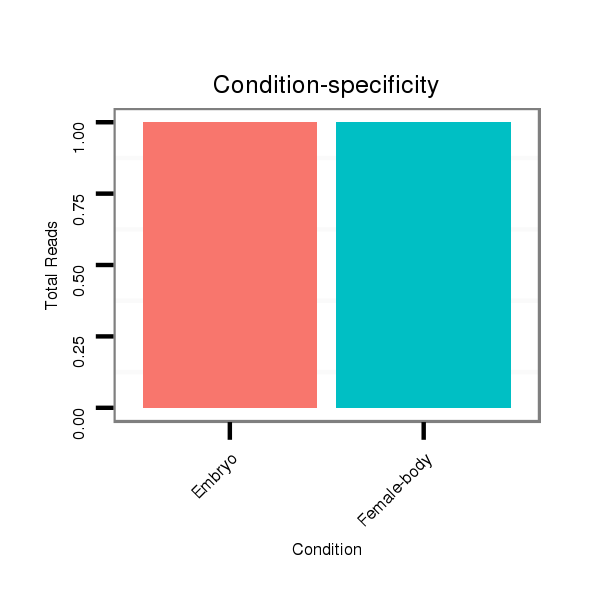
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M061 embryo |
SRR060669 160x9_females_carcasses_total |
SRR060680 9xArg_testes_total |
M027 male body |
V053 head |
V047 embryo |
SRR060660 Argentina_ovaries_total |
SRR060671 9x160_males_carcasses_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................TTCAACAAGTGCAGTGCGATCGATAG..................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GATCTGCGATTGTTTACCATGTA............................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TAACTGAGGCGCATTGTGACTGTCTT................................................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GTTGATATGGCCGAGTTGG........................................................................................................................................................................................................... | 19 | 2 | 6 | 0.50 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TAAAAGATCTGCCATTGTT........................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TCAGAGCTAGCTATGGTTAG........................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................GTTGATATGGCCGAGTTG............................................................................................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AGCCAGGCATGTTTAGGAGA.................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................GTTGAGATGACCGAGTTGG........................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GCAGTGCGCTCGATAGAAA.................................................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............ATTGTGACTGCCTTG................................................................................................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATTGACTCCGCGTAACACTGACAGAACAACAACTATACGGGCTCCACCTCCATTTTTGTCGGGTGTCCCCATAGGCAAGTTGTTCACGTCACGCTAGCTATCTATTGGATTAAACTCTTTGTTAAGAGGTCTCGATCGGTACAAATCTTCTAGACGCTAACAAATGGTACATCTAGTCACTAGGGACGCGACCGTAAAATCGACACTACTCTCTTTCCACGTAGTACGCTTTCTATTCTAGACTTTTCTTC
*******************************************************************************...(((((.((((....((((..((((((..(((.((((((((....)))).))))..)))...))))))...)))))))).))))).........**************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060686 Argx9_0-2h_embryos_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
M028 head |
SRR060672 9x160_females_carcasses_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................ACTGACAGAACAAGAGCTAG....................................................................................................................................................................................................................... | 20 | 3 | 7 | 3.29 | 23 | 23 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................TCGTAACGAGGGACGCGACCGTAA...................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CGAGGGACGCGACCGTAA...................................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AGGGACGCGACCGTAACAG................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................ACTCTCTTTCTACGTAATA......................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............TCACTGTCAGAACCACAAC.......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| ......................................................................................................................................................................................GGGACGCGACCGTAACAG................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................TTGGCAAGTAGTTCAAGTC................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:17155634-17155884 + | dvi_7423 | T---A---ACTGAGGCGCATTGTGACTGTCTTGTTGTTGATATG------CCCGAGGTGGAGGTAAAAACAGCCCAC------------------AGG-----GGTATCCGTTCAACAAGTGCAGTGCGATCGATAGATAACCTAATTTGAGAAACAATTCTCCAGAGCTAGCCATGTT---TAGAAGATCTGCGATTGTTTACCATGTAGATCAGTGATCCCTGCGCTGGCATTTTAGCTGTGATGAGAGAAAGGTGCATCATGCGAAAGATAAGATCTGAAAAGAAG |
| droMoj3 | scaffold_6496:10845851-10846054 - | GGTTAGCTGCTGCGACGCATTGTGACTGTCTCGTAGTTGATATGCCCATGCCCGAGGTGGAGGTAAAAGCAATCCGCGAGCGGATTGTGTAGTAAGCCGAATGGCTTTAGA---GTCGAGTGCAGTGCGATCGAACGAAAGGGAAAGCAGAGAGAAAA-GCTTCTCAGCTACGCGCATTACCTAAGAGATCACGAATTCTTTGTTACG--------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15245:16685012-16685107 + | T---A---GCTCAAAAAGATTGTAACTGTATT---GTTGATATC------CCAGAGACGGAGGTAAAA----------------CTGTGTCACACACC-----CCCTTCGAATAATCGAGTGTAATGCGATC------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 04:18 AM