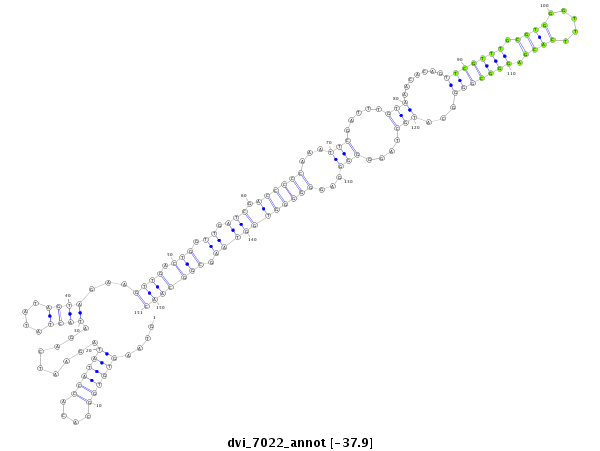
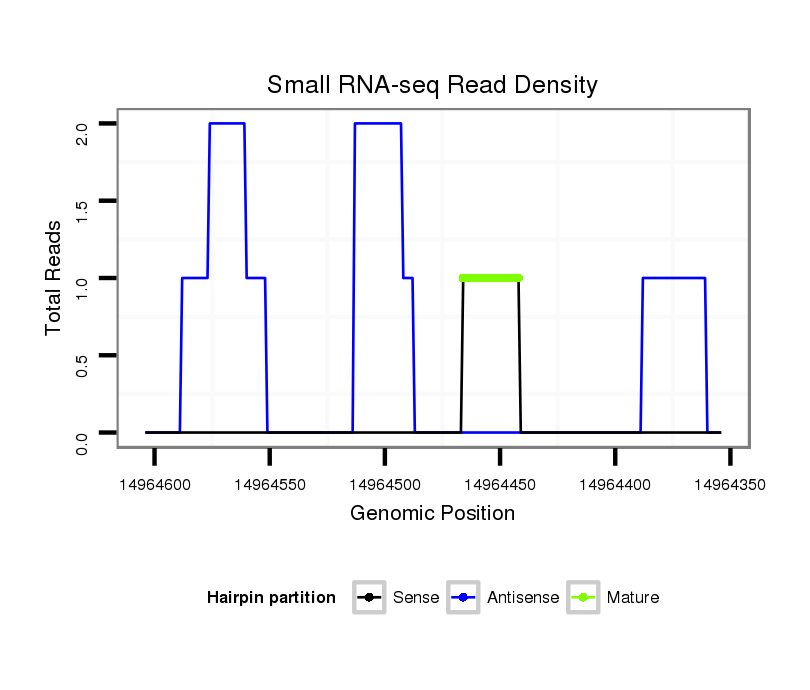
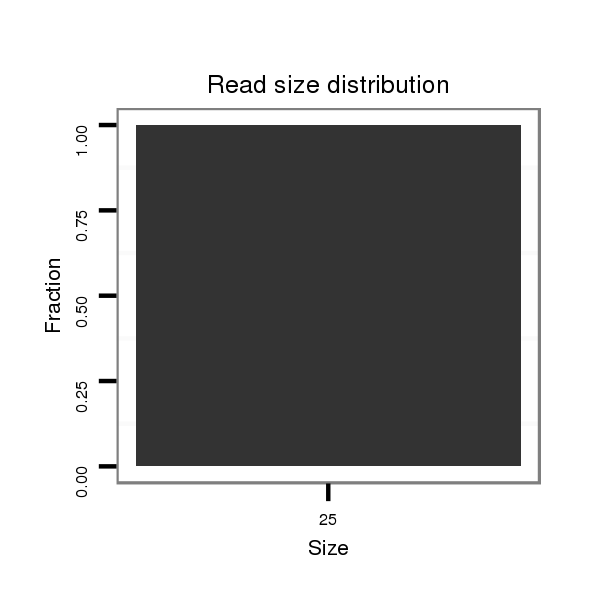
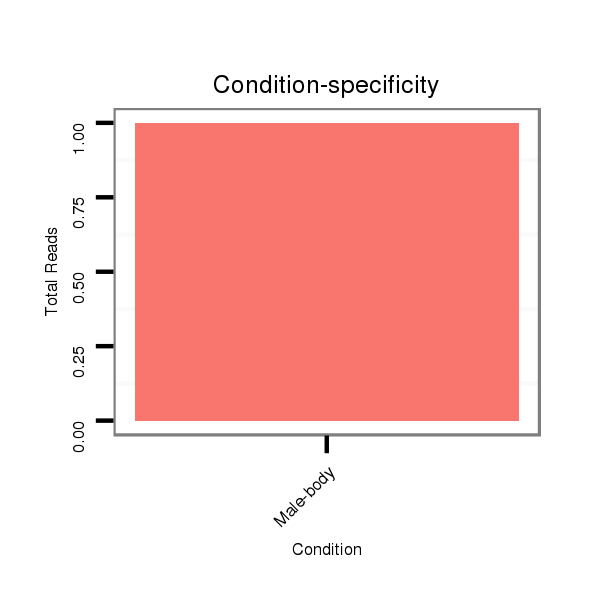
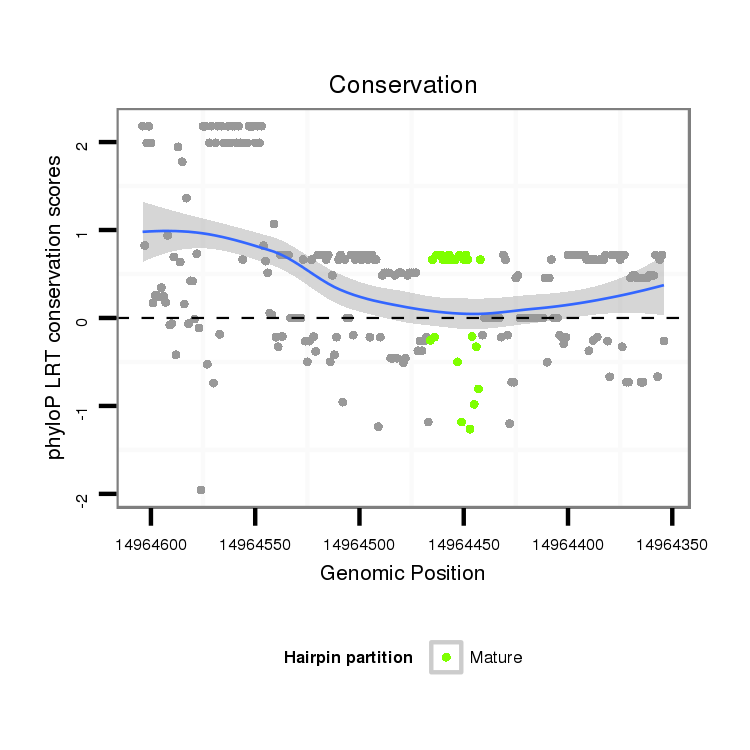
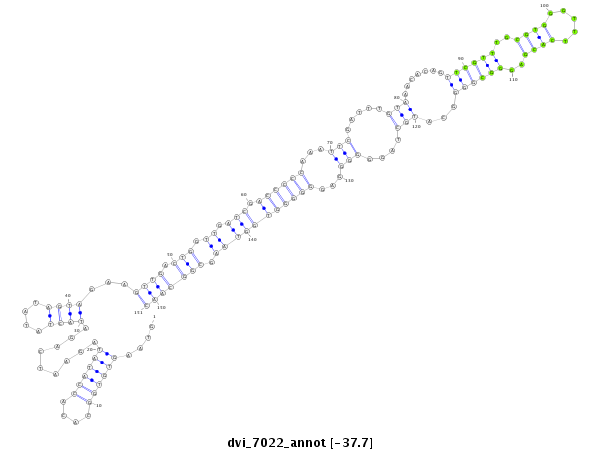
ID:dvi_7022 |
Coordinate:scaffold_12875:14964404-14964554 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.7 | -37.7 | -37.5 |
 |
 |
 |
CDS [Dvir\GJ20215-cds]; exon [dvir_GLEANR_5665:1]; intron [Dvir\GJ20215-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGCAGCCGCTGCTGCAGAGCAAACGCCACATCCTTGACTCCAGATGTGCGGTAAGTGTGGCACACCATATAGAATCAGATACTATATAGTAGAAGTTGACTGGTTGATCGACCCCCAAATTCGATTTGTAAACACAGTTCGTTTGCGTGGGTTTCACGAGGGCGGGGCATGCTAGGGGGGAGGGGGGTGGTAAGCGGCAACCCTATAAATATAGCGTGTGCCAAGAATAGCTGCGATCGCGCCAAATCGAA **************************************************....((((((....)))))).........((((....))))...((((.(((.((.(((.((((((...(((.....(((.......(((((((.((((.....)))).)))))))...)))....)))...))))))))))).)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
V053 head |
V047 embryo |
M061 embryo |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................TCGTTTGCGTGGGTTTCACGAGGGC........................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GAAGGGGGTGGTTAGCGGC..................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................TGCGTGAATTTCACGAGG.......................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................GAATGGTTGGACGACCCCC....................................................................................................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACTACAAGTTGACTGGT................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................CGGGGCAGGATAGGTGGGA...................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TCGTCGGCGACGACGTCTCGTTTGCGGTGTAGGAACTGAGGTCTACACGCCATTCACACCGTGTGGTATATCTTAGTCTATGATATATCATCTTCAACTGACCAACTAGCTGGGGGTTTAAGCTAAACATTTGTGTCAAGCAAACGCACCCAAAGTGCTCCCGCCCCGTACGATCCCCCCTCCCCCCACCATTCGCCGTTGGGATATTTATATCGCACACGGTTCTTATCGACGCTAGCGCGGTTTAGCTT
**************************************************....((((((....)))))).........((((....))))...((((.(((.((.(((.((((((...(((.....(((.......(((((((.((((.....)))).)))))))...)))....)))...))))))))))).)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060676 9xArg_ovaries_total |
SRR060677 Argx9_ovaries_total |
M047 female body |
SRR060654 160x9_ovaries_total |
SRR060656 9x160_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060669 160x9_females_carcasses_total |
SRR060675 140x9_ovaries_total |
V116 male body |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................GTAGGAACTGAGGTCTACACGCCAT...................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGGGATATTTATATCGCGCACGGTT........................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................TAGCTGGGGGTTTAGGCTAAACATT........................................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGGGATATTTATATCGCGCACGGTTC.......................... | 26 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................ACACGGTTCTTATCGACGCTAGCGCGGT....... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGGGATATTTATATCGCACACGGTTCTT........................ | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TTGTGTCGAGCAAACGCAT...................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................CTCGTTTGCGGTGTAGGAACTGAGGTCT............................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................CTTCAACTGACCAACTAGCTG........................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CTTCAACTGACCAACTAGCTGGGGGT...................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................ATATCGACGCTAGCGCGGA....... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................CGGTTGTGGTGTGGGAACTG..................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................TGACCAACAAGCTGGCAGTT..................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:14964354-14964604 - | dvi_7022 | AGCAGCCGCTGCTGCAGAGC---------AAACGCCACATCCTTGACTCCAGATGTGCGGTAAGTGTGGCACACC---ATATAGAATCAGATACTATATAGTAGAAGTTGACTGGTTGATCGACCCCCAAATTCGATTTGTAAACACAG---------------------TTCGTTTGCGTGGGTT--TCACG----AGGGCGGGGCATGCTAGGGGGGAGGGGGGTGGTAAGCGGCAAC-CCTATAAATA---TAG--------------CGTGTGCCAAGAATAGCT----GCGATCGCGCCAA---ATCGAA |
| droMoj3 | scaffold_6496:8877108-8877363 + | AGCAGCCATCGCTGCAAAGC---------AAATGCCGCATCCTTGACTCCAGATGTGCGGTAAGTGTATATCATAGCTATATA------GTGTCTATATAGTC-TAGTCG--TGGTTGATCAACCGT-----------------TTTTATAAACACTTTCGTAAACATTTGCCATTTGCGTGGGGTATGCACCTCCGAATCCG--------TAGGATTGA------------GTGG---C-TCTATAAATACTATAGAGAAGGAGGAAGACTGCGTGCCATTAATAGATCGAATAGATCGATCCAAGATATAGAT | |
| droGri2 | scaffold_15245:14986839-14987069 - | AGCAGCCGATGCTGCAAAGC---------AATCATCGAATCCTTGACTCCAGATGTGCGGTAAGTGTGGCATACC---ACATA------GATACCCTATAGTAGTTGTAG--TAGTTGATCGACCTCCAAACCCAATACGTAAATATAGTAATCGTTACTGTAAACATGCCTCGTTTGCGTGGGTT--CCACTTTCAGTGCCA--------AAGAC-------------------G---TGCAAATAAATA---TAGAG--------AGTCTGTGAGCCAATAATAGCT---------------------TAGAA | |
| droWil2 | scf2_1100000004954:2873074-2873131 - | ATCAGCAGCCACATCA---------------GCGTCTGATCCTTGACTCCAGATGTGCGGTAAGTGTGGCACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | 3:7737649-7737708 + | ATCAGTCGCTTATGCCCAAC---------CAGCGCATCATGCTCGATTCCAGATGTGGGGTAAGTGTGG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_4:1648262-1648321 + | ATCAGTCGCTTATGCCCAAC---------CAGCGCATCATGCTCGATTCCAGATGTGGGGTAAGTGTGG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBip1 | scf7180000396428:470833-470900 + | ATCAGACTCTGATGGCGAGCAACCCGAGCCAGCGCATAATGCTCGATTCCAGATGTGGGGTAAGTGTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302143:584562-584616 + | ATCAGTTGCTGATAGCCAGC---------------AGCATTCTGGACTCCAGATGTGGGGTAAGTGTGGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415666:101755-101809 + | ATCAGTCGCTGATATCCAGC---------------ATTATTCTCGATTCCAGATGTGGGGTAAGTGTGGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409464:409869-409923 + | ATCAGTCGCTGATATCCAGC---------------ATTATACTTGATTCCAGATGTGGGGTAAGTGTGGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 2R:20014301-20014355 + | ATCAGTCGCTGCTCTCCAGC---------------ATGATGCTGGACTCCAGATGTGGGGTAAGTGTGGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 05:15 AM