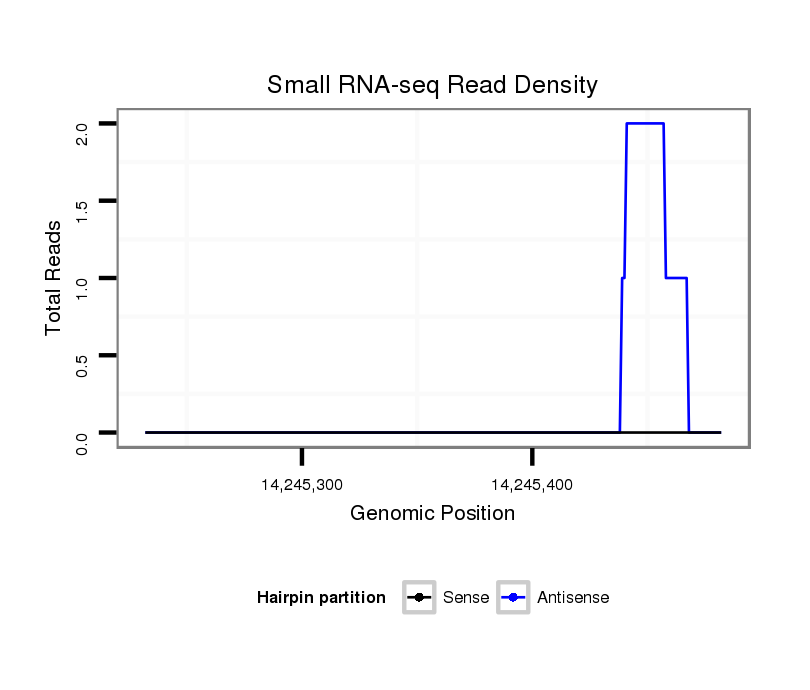
ID:dvi_6836 |
Coordinate:scaffold_12875:14245282-14245432 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ22086-cds]; exon [dvir_GLEANR_7398:1]; intron [Dvir\GJ22086-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| (TG)n | Simple_repeat | Simple_repeat | + |
| -------------------------------------------#######--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGTAGTTAACTAACAAAAAGAAAAAAAAAAAAACAAAAACAAAATGAGCAGTAAGTTTTTCCAGCAACAACAGCAGTTGCCGCCAAAGAAGCGGCAAAAGAAGTGACCACAGATAAAAAAATATAAAAAAAAATATAAATGAAAATCTGTGCGTGTGTGTGTGTGTGTGTTATGCATGGCTACTGCCTATCATATCCAAGTTGAGAGAGTCGCTGGGCCTTCCCTTTGCGATTGCGACTGCGAATGCGACT **************************************************.............((((((...(((.(((((((.......)))))))........(((((((((.......((((........)))).......))))))).))...)))...))).)))((((...(((....)))...)))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
V116 male body |
SRR060668 160x9_males_carcasses_total |
SRR060672 9x160_females_carcasses_total |
M027 male body |
SRR060666 160_males_carcasses_total |
SRR060684 140x9_0-2h_embryos_total |
V053 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................AAGATAAATGAAAGTCTGTGCGTG................................................................................................ | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................AAACAAAAACAAAATGAGC.......................................................................................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CATAGCCGAGTTGAGAGA........................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCATAGCCGAGTTGAGAGA........................................... | 19 | 3 | 20 | 0.25 | 5 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........CTAACAAAAAGAAAAAAAA............................................................................................................................................................................................................................... | 19 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CCGCGAGTGAAGCGGCAAAAG....................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AGATAAATGAAAGTCTGTG.................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CGGCCAGTGAAGCGGCAAAAG....................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TCATAGCCGAGTTGAGAGAC.......................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CGGTGGAGAGAGTCGCTGG................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................TTGCCGCAACAACAGCAGTT............................................................................................................................................................................. | 20 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................CTGCTGTTGCCGCCAAGGAAG................................................................................................................................................................ | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AGAAATATAAAAAAAAAGATGAAT............................................................................................................... | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCATCAATTGATTGTTTTTCTTTTTTTTTTTTTGTTTTTGTTTTACTCGTCATTCAAAAAGGTCGTTGTTGTCGTCAACGGCGGTTTCTTCGCCGTTTTCTTCACTGGTGTCTATTTTTTTATATTTTTTTTTATATTTACTTTTAGACACGCACACACACACACACACAATACGTACCGATGACGGATAGTATAGGTTCAACTCTCTCAGCGACCCGGAAGGGAAACGCTAACGCTGACGCTTACGCTGA
**************************************************.............((((((...(((.(((((((.......)))))))........(((((((((.......((((........)))).......))))))).))...)))...))).)))((((...(((....)))...)))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060688 160_ovaries_total |
V116 male body |
M061 embryo |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................CCGAGGACGGATAGGACAGGT..................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AGCGACCCGGAAGGGAAACGCTAACGC............... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................TCAGCGACCCGGAAGGGAA......................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TACCGAGGACGGATAGGACAGGT..................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTTTTTTGTTTTTGTTTTAC............................................................................................................................................................................................................. | 21 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................AAAAAGGTTGTTGTCGTCGT................................................................................................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:14245232-14245482 + | dvi_6836 | AGTAGTT----AA-CT--AACAAAAAG--------AAA-------------AAAA-AAAAAACAAAAACAAAATGAGCAGTAAGTTTTTC-CAGC-A-----ACAACA-------------------------GC--AG---------T-------TGCCGCCAAAGAAGCGGCAAAA-GAA-------GTGACCACAGATAAAAAAAT--AT-AA-AAAAAAATATAA-AT----------------------------------------GA----AAATCTGTGCGTGTGTGTGTGTGT--------G--------------TGT----TATGCATGGCTACTGCCTAT-CATA-----------TCCAAGTTGAGAGA-GTCGCTGGGCCTTCCCT----------------TTGCGATTGCGACTGCGAATGCG--ACT |
| droMoj3 | scaffold_6496:9700336-9700589 + | ACATATT---TAA-CA------AA-----ATAATAT---ACAATAGTTAAACAAA-ATAAATCAAATACAAAATGAGCAGTAAGTTTTTACCAAC-A-----ACAAGA-------------------------GC--GT---------T-------CGCCACCAAA--------GAA------------GTGAACACAGATAAAAAA----------------------------------------AAAAAAGTTAAATCCA-------AAGA----GAGTCTGTGTGTGTGC------GTGTGTATGTG--------------TGT--ACTATGCATGGCTACTGCCTATCCATA-----------TCCACGTTGAGAGGCGCTGCTGGGCCTTCCCT----------------TTGCAATTGTGTTTGCAATTGCG--ACT | |
| droGri2 | scaffold_15245:14224243-14224455 + | ACTATTT---TAA-CT------AACACAC-----AAAA-------------CAAA-AAATTTCAAAATCAAAATGAGCAGTAAGTTTATC-CAAC-A-----ACAA------------------------------------------CAGAC-------GCCAAA--------AAAATGAA-------GTTATCACAGATAAAAAAAA--AAT---------------AAAAAA------------TATA------AAATCA-------ATGA----GAATCTGTGTGAGTGTGTGTGTGT--GAGTGTG------------TGTGCGTCTTATGCATGTCTGCTG---------G-----------TCCACGT-----------------------------TAT-TGAGT-------------------TGAGGCG--ACT | |
| droWil2 | scf2_1100000004514:136858-137008 - | AACT--T---AAA-CA------AA-----A-------A-------------AAAA-AAAAAAATCAAACACAATGAGCAGTAAGTTTTCG-----------------------------------------------------------------------GCAAA--------AAAGTGAA-------G---AT------GAAAAA----------------------GAAGAA-----------GAA---------------GA-AAACTCA----GTG--------------TGTGTGTGTGTGTGTA---------------------------TCACTGTGTGTGTG-AG--CCT-GTAAGCATTAT-------------------GCATG-----------------------------------------CA--AGT | |
| dp5 | 3:3558089-3558262 + | TAAT--T---TAA-CG------AATTTAA--------A-------------CAAA-AAGAAAACCTAACAAAATGAGCCGTAAGTTTCAA-AATGC----AC------TACTCCCCCGCAACCGTTGATTCGGACGCAGAGTGCAG--CAGCCA-------CC--------AGCAAAAG-GC-------A--AAT------GCGAAT----------------------AAAGAA------------AAGAAA-----------------AAGT----GTG----TG--------TGTGTGT--------G--------------TGT-TTATATGCATGGC------------------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_2:3746811-3746990 + | TAAT--T---TAA-CG------AATTTAA--------A-------------CAAA-AAGAAAACCTAACAAAATGAGCCGTAAGTTTCAA-AATGC----AC------TACTCCCCCGCAACCGTTGATTCGGACGCAGAGTGCAG--CAGCCA-------CC--------AGCAAAAG-GC-------A--AAT------GCGAAT----------------------AAAGAA------------AAGAAA-----------------AAGT----GTG--------------TGTGTGTGTGTGTGTG--------------TGT-TTATATGCATGGC------------------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13266:4861408-4861616 + | dan_1785 | AACT--A---AAA--A------AA-----A----------------------CACTCTAAAAACCTAACAAAATGAGCAGTAAGTTGGGA-TAAAAAAGAAAT----------------------------GAAAGCAAACTGCAA--GCGCCAC-TG----C--------AGCAAAAA-AA-------AAAAAAGCAGAAAAAAAAA--TTA-AATAAAAAAGAATAA-AAGAA------------AAGAGA-----------------AAGAGATGGCG--------------TGTGTGTGTGTATGCA---------------------------TGGCTCAT----AA-AAGG-----------CCCAAAAGGAG----A-----AAGCCTGCCCT-------------------------------------------- |
| droBip1 | scf7180000395751:665339-665505 + | AGTTAAC------CT-TTAATAAA-----A---CACAGA-------------CAC-TCTAAAACCTAACAAAATGAGCAGTAAGTTGGGA-TTAAA-TGAAAT----------------------------GAAAGCAAACTGCAA--GCGCC-------AGCAAA--------AAAAT--------------------------------TA-AA-AGAAAAGAATAA-AAGAA------------AAGA------AAAGTA-------AAGAGACGGCG--------------TGTGTGTGTGTATGCA---------------------------TGGCTA---------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302682:180433-180682 - | ATCGAAAAATTATCC-AAGA--AA-----A-------GACCC----------CAA-GAAAACCCCTAACAAAATGAGCAGTAAGTTGGGA-GGAGC----AC------T------------------------GC--AA-CT--------GCAATTCG----CAAT--AAA--AAAAA----AAGAAAT--G-------ATAA---------------------------AAGAAAAGCGCAAAGC----------------AAACGCAGAAAA----GAGTGTGTGTGCGTGC--------------GTTCGAGCGTGTGTGTGTGT-GCGTATGCATGGCTAAAGGCTGC-AG--GCT-GC--AACCGAA------AAAA-A-----GAGCCTGCCCCAAGTTAAAAGCTC-------------------TAATAAT--CCT | |
| droFic1 | scf7180000453811:192883-193099 - | AGTT--T---A-CCC-ACAATAAA-----A---------CCCCGAGTTAAA-CCC-ATAAAATCCTAACAAAATGAGCAGTAAGTTTTGC-GATAC-CAAAATCAAAAC------------------------GC--AA-AT----GC----------------------------------AAGCAGC--G-------AAAAA--AAAATTA-AA-AAAAAAGA--AA-AAGAA-------AAGCAA-----------A----GC-AAGAAGA----GAG--------------TGTGTGTGTGTATGCA---------------------------TGGCTAAAGGCTGG-AA--ACTTGAAAAACTGAAAATCGAAAGA-A-----AAGCCTGCCCT-------------------------------------------- | |
| droRho1 | scf7180000780219:55821-56047 + | CCAA--A---TAA-AA------AA-----A---------CCCACAGTTAA--TAC-ATAAAATCCTTTCAAAATGAGCAGTAAGTTGGGC-GACAT-CGAAATCCAAA-------------------------GC--AA-AT----GC----------------------------------AAGAAAC--G-------AAAAA--A----------------------AAAGAC-----------AA-----------A----GC-AAGAAGA----GAG--------------TGTGTGTGTGTATGCA---------------------------TGGCTAAAGGCTGG-AA--ACT-GAA-AACTGAAAATCGAAAGA-A-----AAGCCTGCCCTAAGTTAA-AGCTCCTTTGGCGTTTCC---TGCTAATCAACCAAC | |
| droEug1 | scf7180000409474:858283-858521 - | CCAT--T---AAA-AA------AA-----A---------CCCAGAGTTAA--CCC-ATAATATCCTAACAAAATGAGCAGTAAGTTGGGC-GATAC-CGAAAATCAAA-------------------------GCAAAAAATAAA-GC----------------------------------AAGAAAC--G-------AAAAA--A-------------------------AAATA--GAAAAGC----------------AAGC-AAGAGAA----GAG--------------TGTGTGTGTGTATGCA---------------------------TGGCTAAAGGCTGG-AA--ACT-GAACAACTGAAAATCGAAAGA-A-----AAGCCTGCCCTAAGTTAA-AGCTCCTTTGGCGTTTCC---TGCTAATCAACCAAC | |
| droYak3 | 2R:12108072-12108212 - | TCCCCCC---CAC-AA---AA-AAA----A---TATAT--------------CAA-ATAAAATCCTAACAAAATGAGCAGTAAGTTGGGC-CACAA-CCAAATCCAGA-------------------------GC--A--------------------------AA-------CAAATA-AG-------A--AAA------GCAAGA----------------------AAAGAA------------AAGA----------------AAAAAGA----GAG--------------TGTGTGTGTGTATGCA---------------------------TGGCTA---------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/16/2015 at 11:40 PM