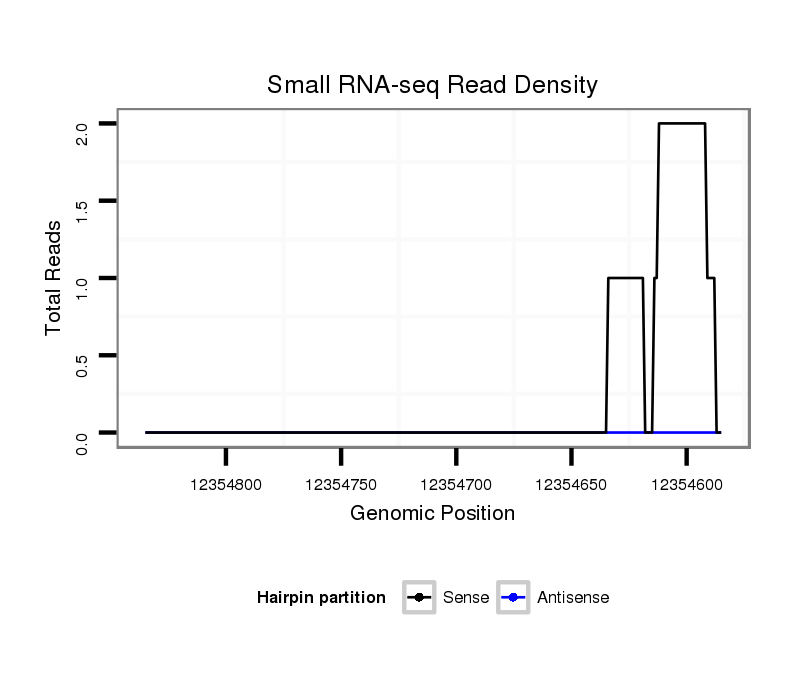
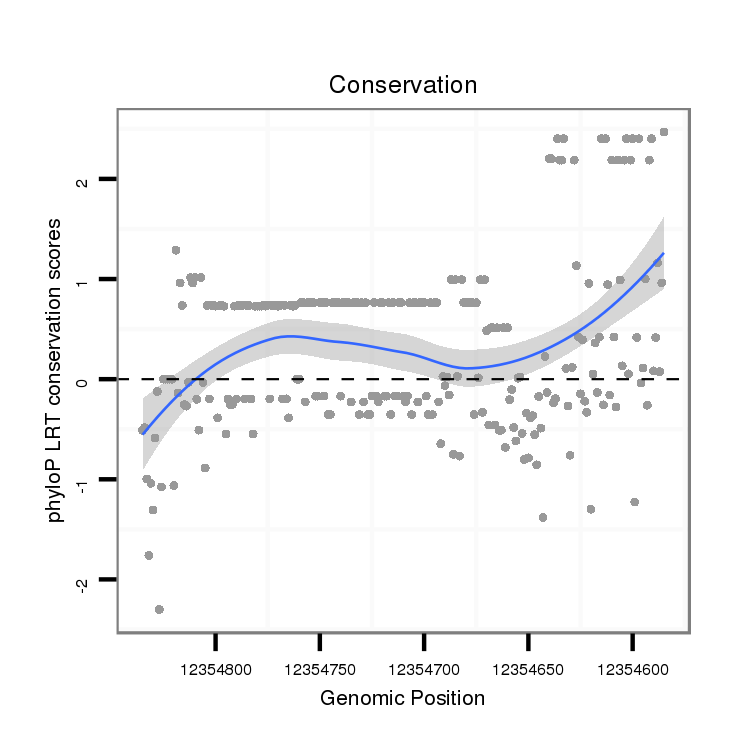
ID:dvi_6454 |
Coordinate:scaffold_12875:12354635-12354785 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ20407-cds]; exon [dvir_GLEANR_5847:3]; intron [Dvir\GJ20407-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTTCAAAATTCTCCGCGCATATCTGAAAAGCATAAGCAAGGGTTGTTAATTTTCGGCGTGTCAAAGACAGCTGTTCTTTAATTTCTTCAGTGTTGATTGGAGGTACAGGGGATTTTGTAGTCGAACACACTCAACTACAGCTTTTGGTTCATTAAAATAAGTAGACTTATTTTACTGTTAATATTAATTTTCCATTTCCAGGACGCTGAGGCGAGAAAACTGAAGCGAGGTCACGTGCTGTCCGAGCTGCT **************************************************...((((.((((...))))))))..........((((((((....))))))))..((((((...(((((((.((......)).)))))))))))))...((.(((((((((....))))))))).))........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M061 embryo |
V116 male body |
SRR060682 9x140_0-2h_embryos_total |
V053 head |
GSM1528803 follicle cells |
V047 embryo |
SRR060662 9x160_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060678 9x140_testes_total |
SRR060671 9x160_males_carcasses_total |
SRR060664 9_males_carcasses_total |
SRR060667 160_females_carcasses_total |
SRR060658 140_ovaries_total |
SRR060660 Argentina_ovaries_total |
SRR060673 9_ovaries_total |
M027 male body |
M028 head |
SRR060661 160x9_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060679 140x9_testes_total |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
SRR060674 9x140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................GACGCTGAGGCGCGAAAGC............................... | 19 | 2 | 1 | 65.00 | 65 | 63 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TGACGCTGAGGCGCGAAAGC............................... | 20 | 3 | 7 | 4.14 | 29 | 16 | 2 | 0 | 5 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGATTAGAGGTTCCGGGG............................................................................................................................................ | 18 | 3 | 14 | 3.50 | 49 | 2 | 0 | 12 | 0 | 0 | 0 | 0 | 5 | 8 | 1 | 4 | 0 | 6 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCGCGAAAGCG.............................. | 20 | 3 | 6 | 2.67 | 16 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGGACGCTGAGGCGCGAAAGC............................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGCTGAGGCGCGAAAGC............................... | 18 | 2 | 8 | 1.13 | 9 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GATTAGAGGTTCCGGGGA........................................................................................................................................... | 18 | 3 | 13 | 1.08 | 14 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................GAAGCGAGGTCACGTGCTGTCCGAGCT... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGATTAGAGGTACCGGGGT........................................................................................................................................... | 19 | 3 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TCACGTGCTGTCCGAGCTGCC | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................AGCGAGGTCACGTGCTGTCCG....... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCGCGAAAG................................ | 18 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCGAGAA.................................. | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCGCGAAAGCGG............................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CTGACGCTGAGGCGCGAAAAC............................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................ACTTCTAATATTAATATTCCAT........................................................ | 22 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TGACGCTGAGGCGCGAAA................................. | 18 | 2 | 5 | 0.40 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGATTGGAGGTTCCGGGGT........................................................................................................................................... | 19 | 3 | 8 | 0.38 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCGAGAAAGCGT............................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGGACGCTGAGGCGCGAAA................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACGCTGAGGCTCGAAAAC............................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TGACGCTGAGGCGTGAAAGC............................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGCTGAGGCTCGAAAAC............................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGATTAGAGGTTCCGGGGA........................................................................................................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TGATTAGAGGTTCGGGGGA........................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACACTGAGGCTCGAAAAC............................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ATTAATATTCCATTTGCAG.................................................. | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................GATTAGAGGTACCGGGGT........................................................................................................................................... | 18 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................ATGATTGGAGGTTCCGGG............................................................................................................................................. | 18 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GTTGATTGGATGAACGGG.............................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACGCTGAGGCGCGAAAGCG.............................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GAAGTTTTAAGAGGCGCGTATAGACTTTTCGTATTCGTTCCCAACAATTAAAAGCCGCACAGTTTCTGTCGACAAGAAATTAAAGAAGTCACAACTAACCTCCATGTCCCCTAAAACATCAGCTTGTGTGAGTTGATGTCGAAAACCAAGTAATTTTATTCATCTGAATAAAATGACAATTATAATTAAAAGGTAAAGGTCCTGCGACTCCGCTCTTTTGACTTCGCTCCAGTGCACGACAGGCTCGACGA
**************************************************...((((.((((...))))))))..........((((((((....))))))))..((((((...(((((((.((......)).)))))))))))))...((.(((((((((....))))))))).))........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
V047 embryo |
SRR060686 Argx9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060663 160_0-2h_embryos_total |
SRR060672 9x160_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................CTGTATAAAAGGACAGTTATAAT................................................................. | 23 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CTGTATAAAAGGACAATCATAAT................................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................GTACACGACAGGCTCGAC.. | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TGTATACAATGACAGTTATAAT................................................................. | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................AAACACCAGCTTGTCTGAAT...................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................AAAAGGTTAAGGTCGTGC.............................................. | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................AAGAAGTCACAACGAAGC....................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..AGGTTTAAGAGGCGCGACT...................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................TGGGTCGACAAGAAATTTAA....................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:12354585-12354835 - | dvi_6454 | CTTCAAAATTCTCCGC-GCATATCTGAAA---AGCATAAGCAAGGGTTGTTAATTTTCGGCGTGTCAAAGACAGCTGTTCTTTAATTTCTTCAGTGTTGATTGGAGG---TACAGGGGATTTTGTAGTCGAACACACTCAACTACAGCTT-TTGGTTCATTAAAATAAGTAGACTTATTTTACTGTTAAT-----------------------------------------------ATTAATTTTCCATTTCCAGGACGCTGAGGCGAGAAAACTGAAGCGAGGTCACGTGCTGTCCGAGCTGCT |
| droMoj3 | scaffold_6496:14515611-14515673 + | TCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATTTTACAGGATGCTGAGGCGAGGAAACTGAAACGAGGTCACGTGCTCACTGAGCTGCT | |
| droGri2 | scaffold_15245:8002210-8002333 + | CTCAACTAGA-----G-GCA-----------------------------------------------------------------------------------------------------------------------------------GTGGCTAATCA-------TAAACCTACTAAAATGCTCTT------------------CTTTTTTTCTCGCT--------------------CCGTAAATTTATAGGATGCTGAGGCGCGCAAATTGAAGCAAGGACACGTACTGTCGGAACTGTT | |
| droWil2 | scf2_1100000004382:1096748-1096806 - | CA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTGCAGGACGCTGAAGCGAGACGGCTGAAACGTGGTCATGTTTTGACAGAGCTTTT | |
| dp5 | 3:112114-112169 - | TT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAGGACGTGGAGGCCCGACGCCTAAAACGTGGTCATGTTCTAACGGAGCTTTT | |
| droPer2 | scaffold_2:301104-301249 - | AATGCACTCA-----T------ATTGAGCTTCAAG-----------------------------------------------------------------------------------------------------------------------------------------------------ACAGACCAAAGAAGCAGTGTTGTTTAGTTTAGCTCACACTCATAATAATTGAATTTCATTGCTGTATTTTAGGACGTGGAGGCCCGACGCCTAAAACGTGGTCATGTTCTAACGGAGCTTTT | |
| droAna3 | scaffold_13266:19436079-19436134 + | TT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAGGATTGTGAGGCTCGTCGTCTTAAGCGAGGACACGTATTAACGGAACTTTT | |
| droBip1 | scf7180000392942:872-951 + | CATAAAAAGG-----G-GAATTTTTGAAAATAAAAACAAGAAAGCAAAGCTAACTTCGGGCGTGTTAAAGATAAATGT--------------------------------------------------------------------------------------------------------------------------------------------------------------TTC--------------------------------------------------------------TAATT | |
| droKik1 | scf7180000301927:4219-4304 + | CTCATTTTAA------------------------T-----------------------------------------------------------------------------------------------------------------------------------------------------ACATAT-----------------------------------------------AAAAAAATTTTTATTTCAGGATCTTGAGTCATTGCGTCTTAAACGGGGTCATGTGCTAACAGAGCTTTT | |
| droFic1 | scf7180000453308:19732-19813 + | TTCGGCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGA-----------------------------------------------ATAACTTCTTCTTTTCCAGGATACAGAGACACGTTGTCTGAAACGTGGTCATGTGCTCAAGGAACTTTT | |
| droEle1 | scf7180000490818:7237-7322 + | TTATATTGTT------------------------T-----------------------------------------------------------------------------------------------------------------------------------------------------CTTAAT-----------------------------------------------TATTTTTTTTCTATTTAAGGATGTCGCGTCTCGACGTTTTAAACGGGACCAAGTACTAAAGGAACTCTT | |
| droRho1 | scf7180000778151:5778-5844 + | TAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTATTTTTTCAGGATAATGAGTCAAGACGTCTTAAACGTGGTCATGTGCTACAGGAACTTTT | |
| droTak1 | scf7180000414238:19628-19704 + | TTATTTTATA-----ATGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATTTTTTTAAGGATGTTGAGCGACGACGCCTTAGACGTGGTCATGTACTAAAGGAACTTTT | |
| droEug1 | scf7180000409741:31537-31724 + | TTAAAAAAAG-----ATGT-------------------------------------------------------------TTAAATTCTTTTATGGTTGGTTGGTGGGCATCCTGTTAACATTACATTTAAATTTAATCTACTGAATCTATTCTACTTACGAAAATACGAAAA-----------ACGAATTAAAAAAGC-----------------------------------------TTAAACCTTTTTTCAGGATGCTGAACTACGTTTTCTGAGGCATGGTCATGTACTAAAGGAAATTTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/17/2015 at 04:54 AM