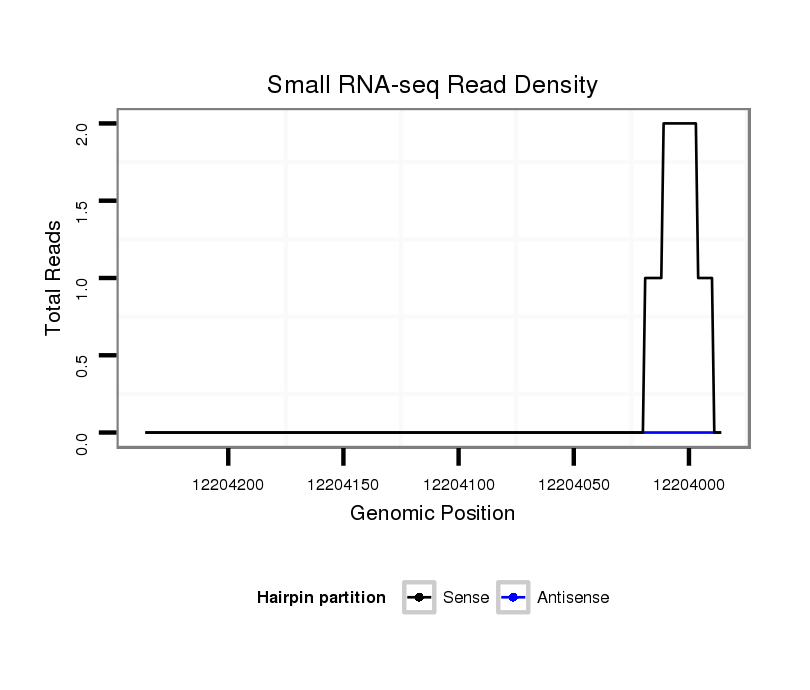
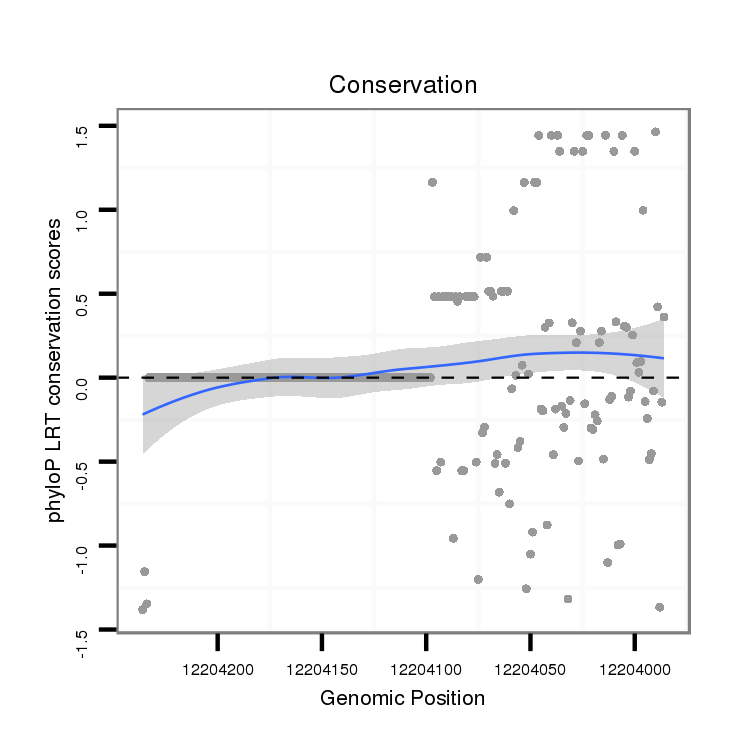
ID:dvi_6418 |
Coordinate:scaffold_12875:12204036-12204186 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ20426-cds]; exon [dvir_GLEANR_5866:7]; intron [Dvir\GJ20426-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCCACTTGCTGCGTACTTGTGGTTAGGTCATTTACGCAAAGGAGGTACTATCTATACTTTGATCGATATTTTTATTTTTATAAAGAAAAAATGTCTTAATTACTACGTTACCATGCACAGTTTTCTTTTAAAGTATTTTTAAACATAAACTGTATAAAATACTGCTTAGAAATAATCTATTATTATGATTTACCGTTCTAGGTTTTACCTTCATATTCACAAACACCACATGGCAACAATGTAACTCAACG **************************************************..............(((((((((.((((...)))).))))))))).....................(((((((...((((((.....))))))...)))))))............((((((..((((.........)))).....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
SRR060667 160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
V053 head |
SRR060668 160x9_males_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR1106726 embryo_14-16h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................CACAAACACCACATGGCAACAAT........... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CCACATGGCAACAATGTAACTC.... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ATGATGATTTACCATTCTAGT................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GTATTCTTAACCATAAACT.................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GCTAGGTTTGACCGTCATA.................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................TGTACGATCTATACTTCGA............................................................................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................CGTAGTCTTAAACATAAACT.................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................ACTACGTTACCGTGG....................................................................................................................................... | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................GATCGTTTTAGGTTTTACC.......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................TAAAGAAAGAATGTCGGAA........................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
CGGTGAACGACGCATGAACACCAATCCAGTAAATGCGTTTCCTCCATGATAGATATGAAACTAGCTATAAAAATAAAAATATTTCTTTTTTACAGAATTAATGATGCAATGGTACGTGTCAAAAGAAAATTTCATAAAAATTTGTATTTGACATATTTTATGACGAATCTTTATTAGATAATAATACTAAATGGCAAGATCCAAAATGGAAGTATAAGTGTTTGTGGTGTACCGTTGTTACATTGAGTTGC
**************************************************..............(((((((((.((((...)))).))))))))).....................(((((((...((((((.....))))))...)))))))............((((((..((((.........)))).....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
V047 embryo |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060687 9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR060654 160x9_ovaries_total |
SRR060675 140x9_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......CGACGCAGGAACGCCAAGCCAG.............................................................................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CCGACGCAGGAACGCCAATC................................................................................................................................................................................................................................. | 20 | 3 | 8 | 0.63 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AAATTTGTGTGTGACATAT............................................................................................... | 19 | 2 | 5 | 0.60 | 3 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| ..........CGCATGAACACCAATCGAGTGC........................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AAAAATGTGTGTGTGACATAT............................................................................................... | 21 | 3 | 18 | 0.17 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 |
| .......CGACGCAGGAACGCCAAGCC................................................................................................................................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....GCCGACGCATGAACTCCAAT.................................................................................................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CGACGCAGGAACGCCAAT.................................................................................................................................................................................................................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TAAATGGGAAGATCTAGAAT............................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......ACGACGCAGGAACTCCAAG.................................................................................................................................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:12203986-12204236 - | dvi_6418 | GCCACTTGCTGCGTACTTGTGGTTAGGTCATTTACGCAAAGGAGGTACTATCTATACTTTGATCGATATTTTTATTTTTATAAAGAAAAAATGTCTTAATTACTACGTTACCATGCACAGTTTTCTTTTAAAGTATTTTTAAACATAAACTGTATAAAATAC--------TGCTTAGAAATAATCTATTATTATGATTTACCGTTCTAGGTTTTACCTTCATATTCA---CAAACACCACATGGCAACAATGTAACTCAACG |
| droMoj3 | scaffold_6496:14656603-14656710 + | TTG----------------------------------------------------------------------------------------------------------------------------------------TATATATAAAGTGTTAAAAATGTTTACTTACTTCT---------------TATGTTCTTTATCTCTTTAGGTGCAACCTTCGTACCAG---CAAATACCAGCTAGCAACAATGCAATTCAGCG | |
| droGri2 | scaffold_15245:12006401-12006494 + | AAT----------------------------------------------------------------------------------------------------------------------------------------T---------------------G--------TGATTAGTGCTATTATATAATTCCATTTTGTCATTTCAGTCTGTACCCTCATACTCA---CACAAGTCACATGGCAACAATGTTCATCCACG | |
| droWil2 | scf2_1100000004513:4968845-4968910 + | TT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTCATTACAGGTTATACAACCATATATAGGAAGTAAACCATCTGGTGACTCCGTTCCCCATAG | |
| droBia1 | scf7180000302143:512960-512966 + | CT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACCG | |
| droEug1 | scf7180000409464:350210-350284 + | TAA----------------------------------------------------------------------------------------------------------------------------------------T--------------------------------------------TGAATTTTTTTTTTTACTGTTTTAGAGATCTCCCTCTTATTCC---CAAAGGTCGAGTGAATCCT---C-TCACAGAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/17/2015 at 04:33 AM