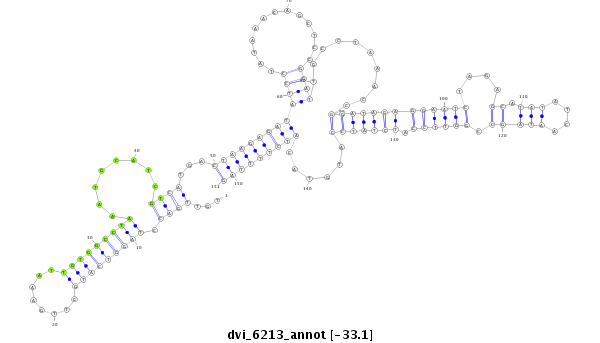
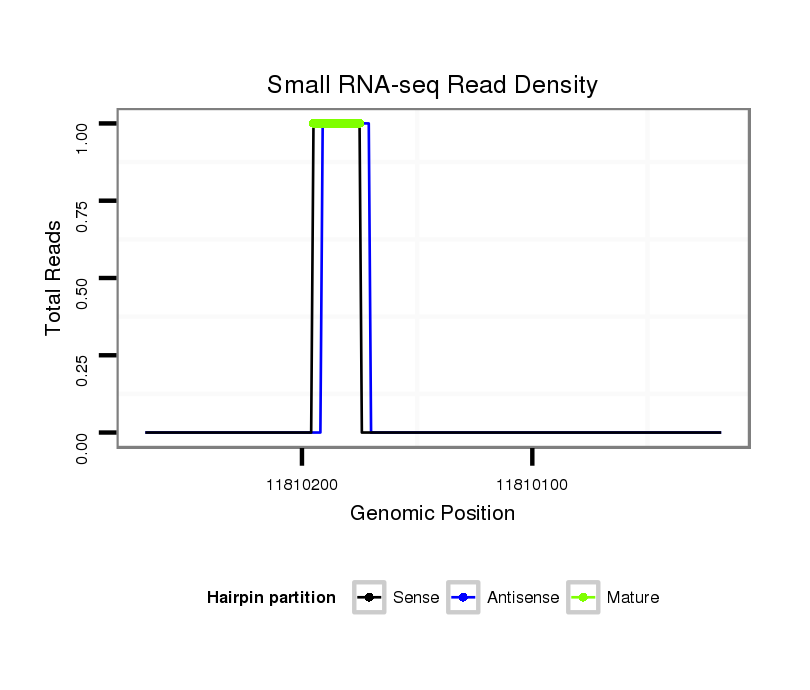
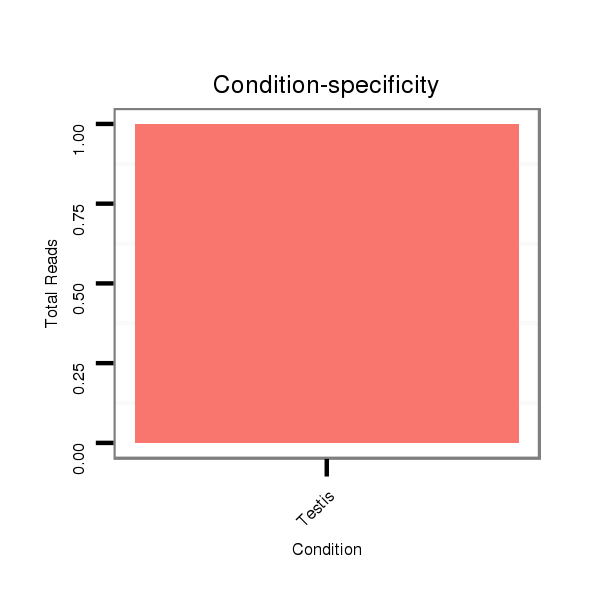
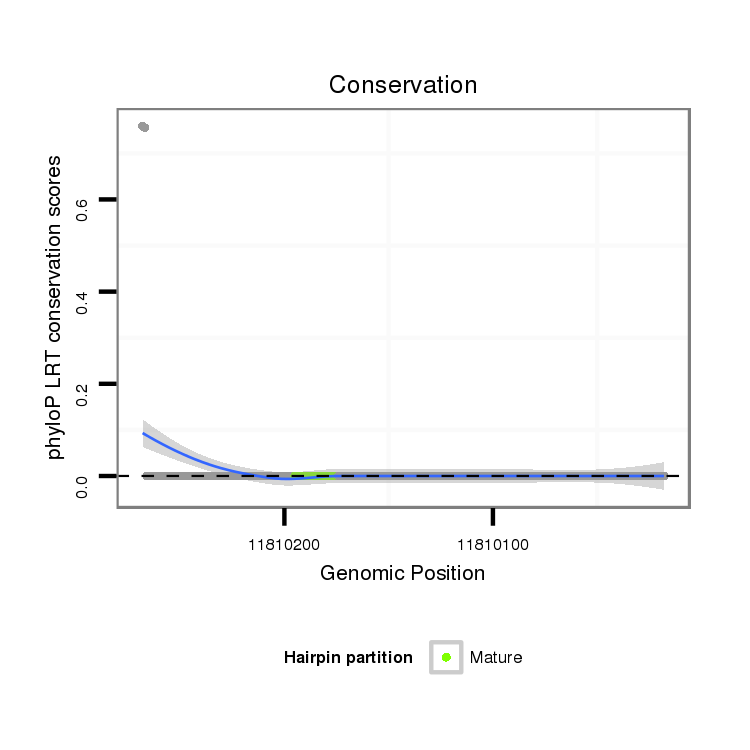
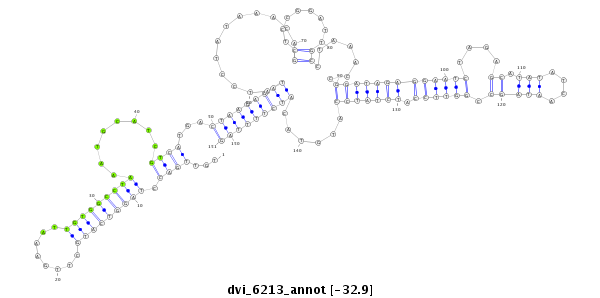
ID:dvi_6213 |
Coordinate:scaffold_12875:11810068-11810218 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.9 | -32.7 | -32.5 |
 |
 |
 |
CDS [Dvir\GJ20470-cds]; exon [dvir_GLEANR_5905:2]; intron [Dvir\GJ20470-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------####################################-------------- TCTACTCGATTGGAATCGGGCGAAGGGGTTCCTATATGAATTAAGTGTAATGTTGACCTAGGTCATGCTTGAAATTGTGGCCTAAATGCATCGTCATGACTAAGAGATATCCTATAAACAGCTCCGGATTGCCTAAACCGGATAGAGGAATCTAGAGCATATATCAATAGCCGGTTCCATCTATCCATGTACATCTTTTAGAGATGGACGCGGGCCATCTCCCGTGCCCCTCAGTAAATATAATACTATGT **************************************************...((((.(((((((((........)))))))))........))))...(((((((((((((.............))))..........(((((((((((((....((.(((....))))).)))))).)))))))......)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V053 head |
SRR060682 9x140_0-2h_embryos_total |
SRR060689 160x9_testes_total |
V116 male body |
M047 female body |
SRR060665 9_females_carcasses_total |
V047 embryo |
SRR1106728 larvae |
SRR060687 9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
M028 head |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................GGCCGAAGGGGTTCATAT........................................................................................................................................................................................................................ | 18 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TTGACTTAACCGGATAGAGGAT..................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................ATTGTGGCCTAAATGCATCGT............................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CAGGTCATCCTTGATATTGTG............................................................................................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CATCGGGAGAAGGGGTGCC........................................................................................................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ATGTCATGCTTGGAATTGTGT........................................................................................................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CATCGTAATGATTAAGGGA................................................................................................................................................ | 19 | 3 | 18 | 0.11 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................GCCCAAATGCCTCGTCAT.......................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TCGGGAGAAGGGGTGC............................................................................................................................................................................................................................ | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................GGCCGAAGGGGTTCATATT....................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............CTTCGGGAGAAGGGGTTC............................................................................................................................................................................................................................ | 18 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TTTCATGCTTGAAATTGTG............................................................................................................................................................................ | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................CATGAAAGTGTGGCCTATA..................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................GTAATGGTGGTCTAGGTC........................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
AGATGAGCTAACCTTAGCCCGCTTCCCCAAGGATATACTTAATTCACATTACAACTGGATCCAGTACGAACTTTAACACCGGATTTACGTAGCAGTACTGATTCTCTATAGGATATTTGTCGAGGCCTAACGGATTTGGCCTATCTCCTTAGATCTCGTATATAGTTATCGGCCAAGGTAGATAGGTACATGTAGAAAATCTCTACCTGCGCCCGGTAGAGGGCACGGGGAGTCATTTATATTATGATACA
**************************************************...((((.(((((((((........)))))))))........))))...(((((((((((((.............))))..........(((((((((((((....((.(((....))))).)))))).)))))))......)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M027 male body |
M047 female body |
SRR060666 160_males_carcasses_total |
GSM1528803 follicle cells |
SRR060665 9_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................CCCGCTTCCCCGAGGATGTA...................................................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................ACCGGATTTACGTAGCAGTAC......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CGGCTTCCCAAAGGCTATAC..................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GGCCTATCTCCTTAGCTT................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................CTTCCCAAAGGCTATACTG................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................AGGCCTAATGAATTTAGCC.............................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................CTATTTGTCGTGGCCTAGC........................................................................................................................ | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AAAGTAGAAAGGTACATG........................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:11810018-11810268 - | dvi_6213 | TCTACTCGATTGGAATCGGGCGAAGGGGTTCCTATATGAATTAAGTGTAATGTTGACCTAGGTCATGCTTGAAATTGTGGCCTAAATGCATCGTCATGACTAAGAGATATCCTATAAACAGCTCCGGATTGCCTAAACCGGATAGAGGAATCTAGAGCATATATCAATAGCCGGTTCCATCTATCCATGTACATCTTTTAGAGATGGACGCGGGCCATCTCCCGTGCCCCTCAGTAAATATAATACTATGT |
| droSec2 | scaffold_376:12232-12233 + | TC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/16/2015 at 09:31 PM