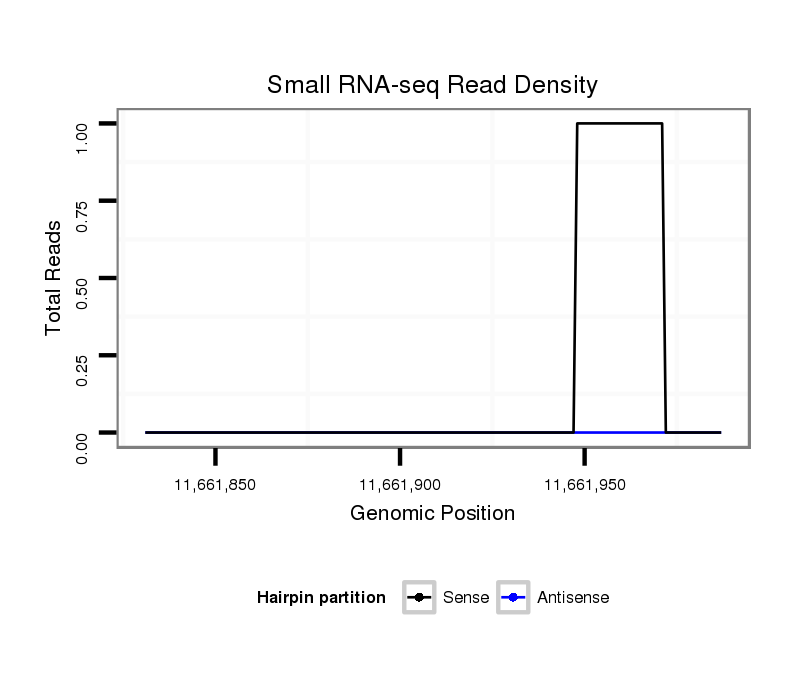
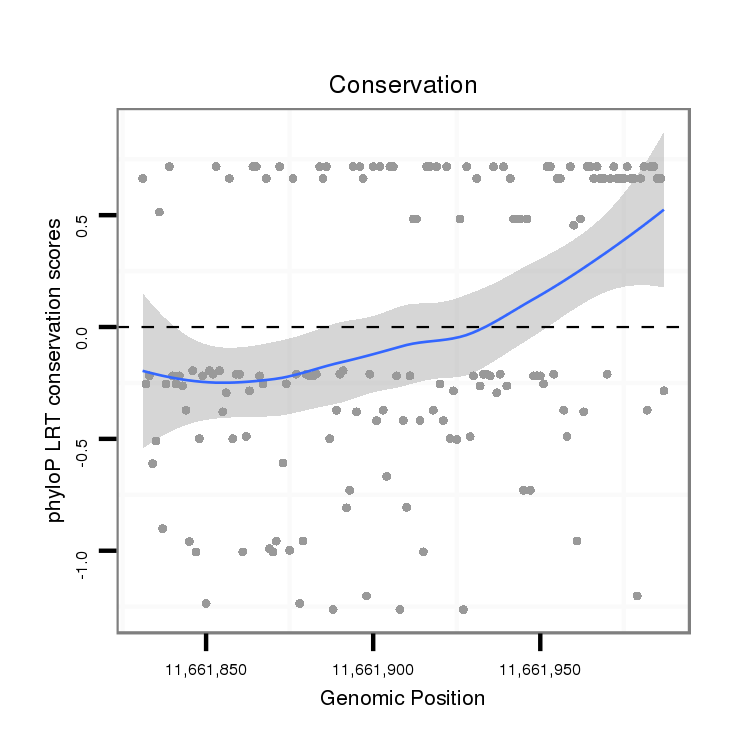
ID:dvi_6165 |
Coordinate:scaffold_12875:11661881-11661937 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ21848-cds]; exon [dvir_GLEANR_7177:1]; CDS [Dvir\GJ21848-cds]; exon [dvir_GLEANR_7177:2]; intron [Dvir\GJ21848-in]
No Repeatable elements found
| ##################################################---------------------------------------------------------################################################## GTTCTTGTAGTGTCACTAGCCATGAGGAAATCAAAGAGTTTAGTCCTGTAGTAAGTAGCTGGTTAAGGTAAACCTTTCTCAAAATAACATTTTACTGTTTGATTTAGAAAGAAACATTGGTAATCGCTAGCAAAACACCAGAGCCACGGCACTTGGT **************************************************...(((((...(((((((....))).........))))...)))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060687 9_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
V053 head |
SRR060670 9_testes_total |
SRR060688 160_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................TGGTAATCGCTAGCAAAACACCAG................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| GATCTTGTCGTGCCACTAGC......................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................ATAGAGTTTAGTGCTGTA........................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................GCCATGAGGGAAGCAAAAA........................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................TACTCTCTAGCGAAACACC.................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
CAAGAACATCACAGTGATCGGTACTCCTTTAGTTTCTCAAATCAGGACATCATTCATCGACCAATTCCATTTGGAAAGAGTTTTATTGTAAAATGACAAACTAAATCTTTCTTTGTAACCATTAGCGATCGTTTTGTGGTCTCGGTGCCGTGAACCA
**************************************************...(((((...(((((((....))).........))))...)))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060657 140_testes_total |
SRR060666 160_males_carcasses_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................TGGTCGCAGTGCCATGAAC.. | 19 | 3 | 12 | 0.17 | 2 | 1 | 1 | 0 |
| ...................................CTATAATCAGGACATCA......................................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:11661831-11661987 + | dvi_6165 | GTTCTTGTAGTGTCACTAGCCATGAGGAAATCAAAGAGTTTAGTCCTGTAGTAAGTAGCTGGTTAAG---------GTAAACCTTTCTCAAAATAACATTT--------TACTGTT------TGATTTAGAAAGAAACATTGGTAATCGCTAGCAAAACACCAGAGCCACGGCACTTGGT |
| droMoj3 | scaffold_6496:15206931-15207094 - | GCTG---CAACAATTCGCAGCATGAGGCAAGCGAAAGGAGCACCACTCCAATAAGTCCTTGCGTAAGTAGATATCTATATATATTTGAGAAATGAATACATA-------GGTTCTG------TGTTTTAGAATGAAAAAGTAACAATCGTGAGGAAAACACCAGAGCCACGACATTTGGC | |
| droGri2 | scaffold_15245:6566505-6566670 + | GTACATCTAGTGTTGTAAGTTGTACTGAGGAGGAAGAGGAAAGTTCCAAGGAGAGTATTCAC-TCAGTAAGTGCCACCATATATTAAAGT--TAAATATATTATAATTATG--TTGTCCTTCAGACCAATGAAG------AGGTAACCGTGA---CAACACCGGAGCCACGCCATTTGGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:50 PM