
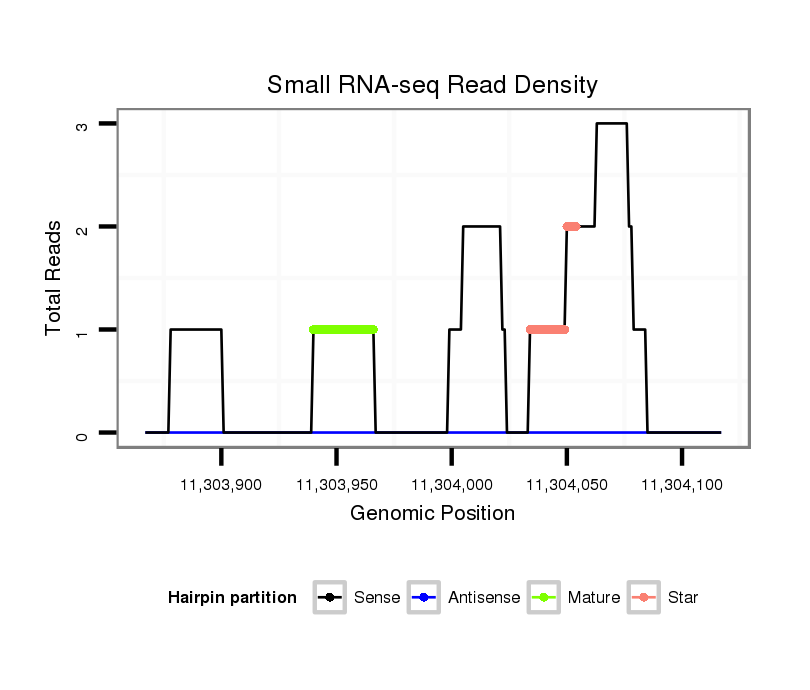
ID:dvi_6101 |
Coordinate:scaffold_12875:11303917-11304067 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.0 | -18.5 | -18.4 |
 |
 |
 |
CDS [Dvir\HP2-cds]; exon [dvir_GLEANR_7152:8]; intron [Dvir\HP2-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCAACAAATTGACGAGGAAATGGGTGATGATTCAGGTGAAAAACCTCTAATGACAAACCCCATTACAAATATCCCAGCTGAAAAGAACCATTTAAAAGCAAATAAGTCAGATAATCCGGAAAACACAATGCAAACGCGACGCTCTGCATTGGCCAAGGCTAGCCCTTGTCCAGGGAATATGTCTCTCGTAATATCGACAGGTCGGGAGAGTCGTAGCAGTAAGCGATTGCTTAGAGGCGACAATAACGCG *************************************************************************....(((..(((......)))...))).........(((((.((((((.....(((((((...((...))..)))))))..(((((.....))))))))..)))...)))))...*************************************************************** |
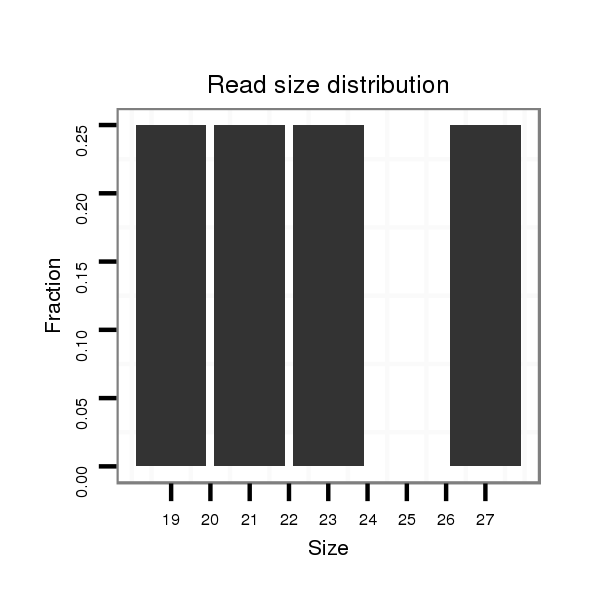
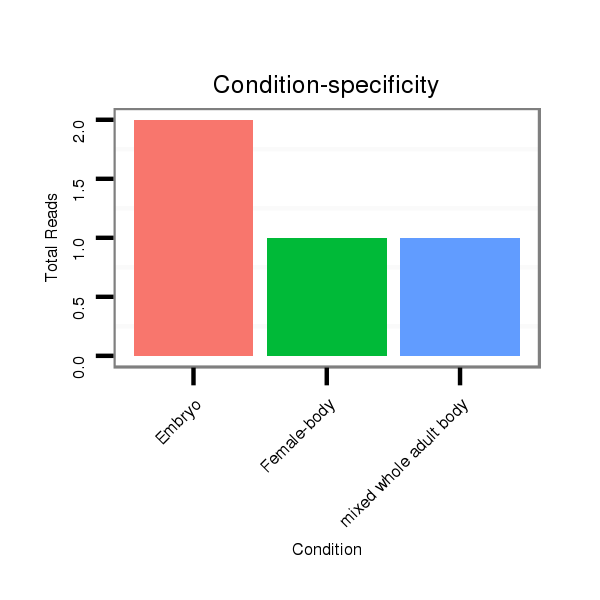
Read size | # Mismatch | Hit Count | Total Norm | Total | M047 female body |
M061 embryo |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
V116 male body |
SRR060667 160_females_carcasses_total |
SRR060663 160_0-2h_embryos_total |
SRR060669 160x9_females_carcasses_total |
M027 male body |
SRR060657 140_testes_total |
SRR060674 9x140_ovaries_total |
V047 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........GACGAGGAAATGGGTGATGATTC......................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................CCCAGCTGAAAAGAACCATTTAAAAGC....................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAAAAGAACCATTTAAAAGCAC..................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TGTCCAGGGAATATGTCTCTC............................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GACGCTCTGCATTGGCCAA.............................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GTAATATCGACAGGTCGGGAGAGT....................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GACAGGTCGGGAGAGTCGTAGC................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTCTCGTAATATCGACAGGTCGGGAGA......................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AAACGCGACGCTCTGCATTGGCC................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................ATGGGTGAGGATTGAGCTGA................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........ATTCACGAAGAAATGGGT................................................................................................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CAAGCGACTGCTTAGAGG............. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AATTAGTCAGAGAATCCG.................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GTAAGCGAATGATTAGAGGC............ | 20 | 2 | 14 | 0.14 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| .................................................................ACAGATATCCGGGCTGAAAA...................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................CCAGCAGAAAAGAACAAGTTA............................................................................................................................................................ | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GTAAGCGAATGATTAGAAGC............ | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................ATGGGTGAGGATTAAGCTG.................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACGTTGTTTAACTGCTCCTTTACCCACTACTAAGTCCACTTTTTGGAGATTACTGTTTGGGGTAATGTTTATAGGGTCGACTTTTCTTGGTAAATTTTCGTTTATTCAGTCTATTAGGCCTTTTGTGTTACGTTTGCGCTGCGAGACGTAACCGGTTCCGATCGGGAACAGGTCCCTTATACAGAGAGCATTATAGCTGTCCAGCCCTCTCAGCATCGTCATTCGCTAACGAATCTCCGCTGTTATTGCGC
***************************************************************....(((..(((......)))...))).........(((((.((((((.....(((((((...((...))..)))))))..(((((.....))))))))..)))...)))))...************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
V116 male body |
SRR060685 9xArg_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060681 Argx9_testes_total |
V053 head |
SRR060661 160x9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060678 9x140_testes_total |
SRR060680 9xArg_testes_total |
M061 embryo |
SRR060679 140x9_testes_total |
V047 embryo |
SRR060676 9xArg_ovaries_total |
SRR060674 9x140_ovaries_total |
SRR060677 Argx9_ovaries_total |
M027 male body |
M028 head |
GSM1528803 follicle cells |
SRR060663 160_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................GAGAGCATGATTCCTGTCCAG............................................... | 21 | 3 | 1 | 58.00 | 58 | 11 | 18 | 6 | 7 | 7 | 0 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATGATTCCTGTCC................................................. | 19 | 3 | 2 | 43.00 | 86 | 31 | 3 | 17 | 13 | 2 | 4 | 7 | 4 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATGATTCCTGTCCA................................................ | 20 | 3 | 1 | 38.00 | 38 | 11 | 1 | 6 | 8 | 1 | 7 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGAGCATGATTCCTGTCCAG............................................... | 20 | 3 | 2 | 33.50 | 67 | 23 | 15 | 8 | 9 | 0 | 2 | 6 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATGATTCCTGTC.................................................. | 18 | 3 | 12 | 11.00 | 132 | 51 | 6 | 36 | 15 | 1 | 5 | 12 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................GAGCATGATTCCTGTCCAG............................................... | 19 | 3 | 2 | 5.00 | 10 | 3 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGAGCATGATTCCTGTCCA................................................ | 19 | 3 | 5 | 2.60 | 13 | 8 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGAGCATGATTCCTGTCC................................................. | 18 | 3 | 19 | 1.32 | 25 | 13 | 0 | 4 | 1 | 0 | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATGATCCCTGTCCA................................................ | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATTATTCCTGTCCAG............................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................GGAGAGCATTATTCCTGTCCA................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATTATTCCTGTCCAGG.............................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGAGCATGATTACTGTCCAG............................................... | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................GAGCATGATTCCTGTCCA................................................ | 18 | 3 | 15 | 0.27 | 4 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................CGATTCTCCGCTTTTATGGC.. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GAGAGCATTATTCCTGTCT................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ACAGCCGTCTAAGCATCG................................. | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGTAAATTGCGGTTTATTC................................................................................................................................................ | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................ACATTTGCTCTGCGAGATG....................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
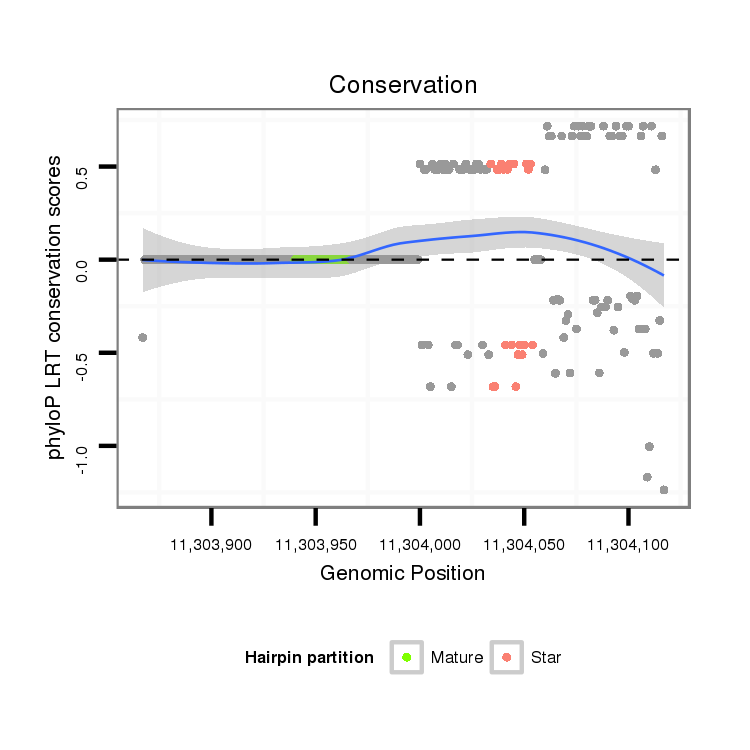
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:11303867-11304117 + | dvi_6101 | TGCAACAAATTGACGAGGAAATGGGTGATGATTCAGGTGAAAAACCTCTAATGACAAACCCCATTACAAATATCCCAGCTGAAAAGAACCATTTAAAAGCAAATAAGTCAGATAATCCGGAAAACACAATGCAAACGCGACGCTCTGCATTGGCCAAGGCTAGCCCTTGTCCAGGGAATATGTCTCTCGTAATATCGACAGGTCGGGAGAGTCGTAGCAGTAAGCGATTGCTTAGAGGCGACAATAACGCG |
| droMoj3 | scaffold_6496:15620112-15620171 - | A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATCGAGAAGAAGCGAAAGTCGTAATGCCAGACGATCGCGTAGAAGTGATGCTGATTCC | |
| droGri2 | scaffold_15245:13425146-13425255 - | A------------------------------------------------------------------------------------------------------------------------------------AGCGTTACGCTCTGCCTCAGCCATGGCTAGTCCATTGCCAGAGAGTCAAATTCTT------TCGTCGGGACTGGAAAGTCGTAGCGGTAAGCGCTTGCTTAAGGATGATCTT---GCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:05 PM