
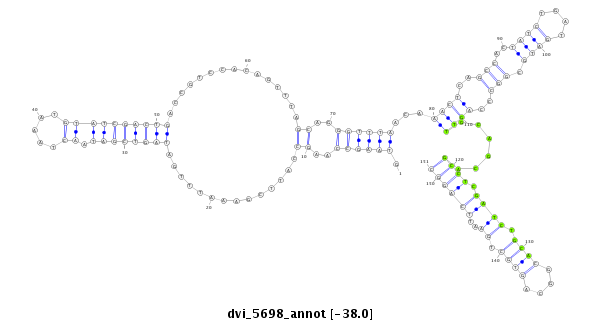
ID:dvi_5698 |
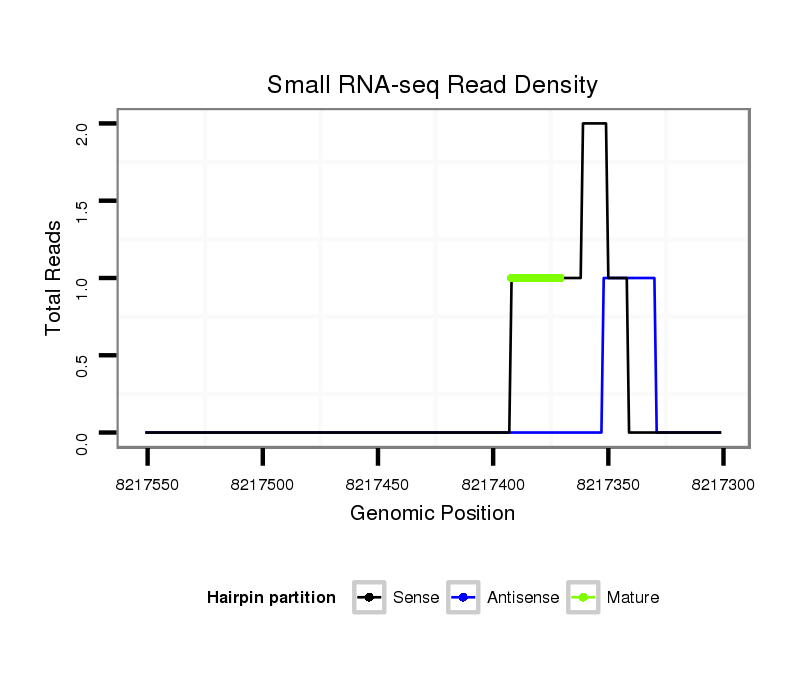
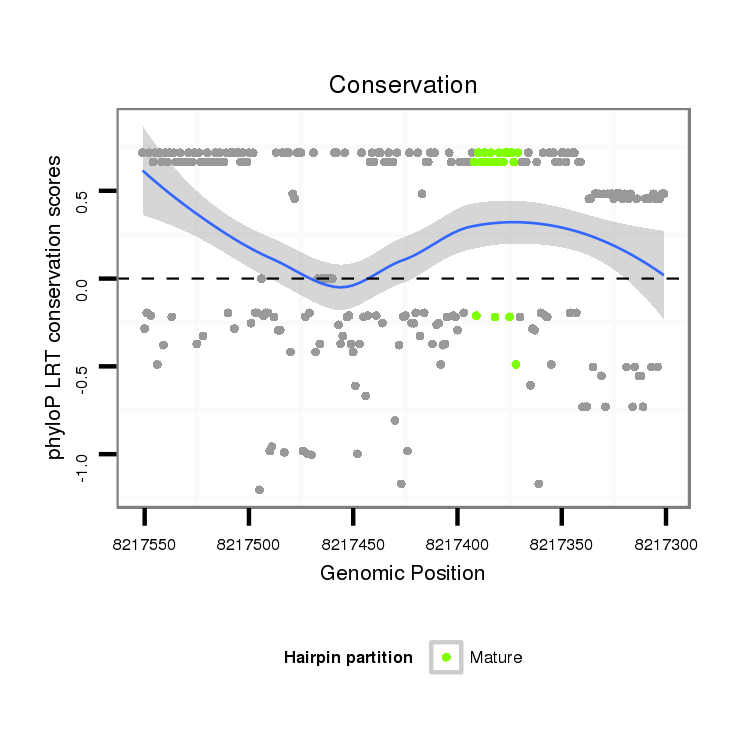
Coordinate:scaffold_12875:8217351-8217501 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -38.0 | -37.9 | -37.9 |
 |
 |
 |
CDS [Dvir\GJ20698-cds]; exon [dvir_GLEANR_6117:1]; intron [Dvir\GJ20698-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AACATCAGTGAACACAGCTCGCAGCTGACGGAGGCACTCGACTTAAAGTGGTAAGCCAAGCCATTCGAAATTTGATAGTCGATAACTAAATGTATCGACTGACCGTCCACAGTTTAGCAGGGTTTAACAAACTCAGCCACTATCTGATGATGCGGCCCAGTTGCAGCAGCCTCGATCTGCACGGCAGTGCTGAATTCAGGCAACTCTAGGCAACGCGAAAATTATTCATAGAACTTACTTCCGGCTCACAA **************************************************.((((((..((..............(((((((((........)))))))))...............))..))))))...((((..(((..((((....)))).)))..))))......((((.((((.((((....)))).))..))))))************************************************** |
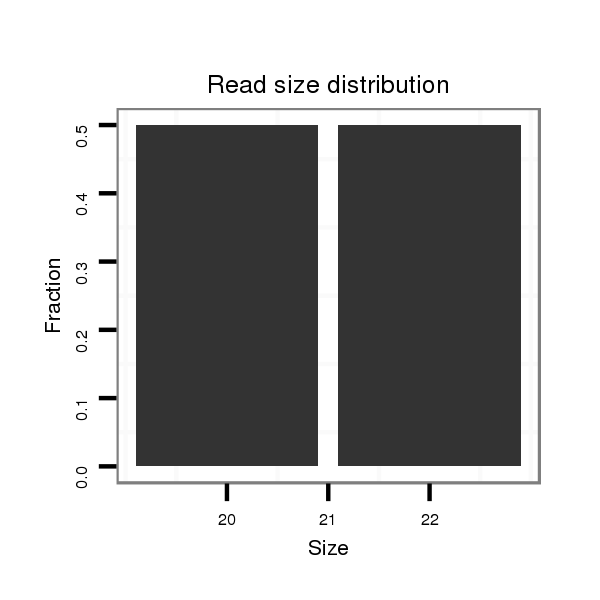
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
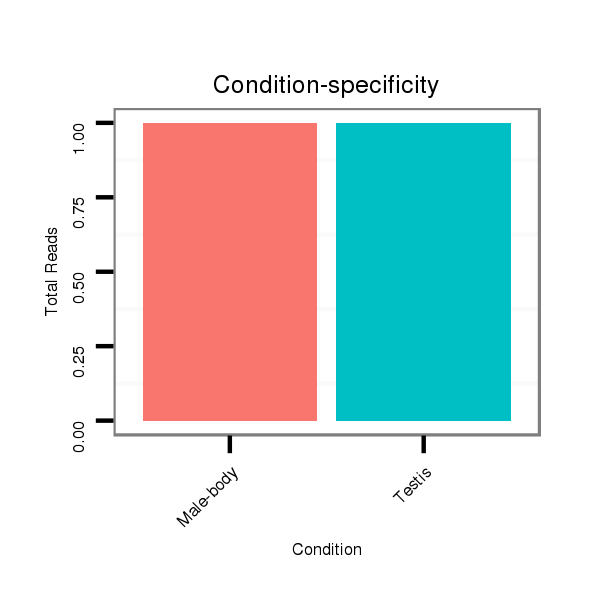
M027 male body |
M047 female body |
V053 head |
|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................GTTGCAGCAGCCTCGATCTGCA...................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CGGCAGTGCTGAATTCAGGC.................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................TGAATTCAGGCAACTCTAGG......................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................TAACGAACTCTGCCACTACC........................................................................................................... | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 |
| ........................................................................................................GTCGACAGTTTATCAGGGTTT.............................................................................................................................. | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................CGGACCGTCGACAGTTTTGC..................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................TCTGCTGGGCAGTGGTGAAT........................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
|
TTGTAGTCACTTGTGTCGAGCGTCGACTGCCTCCGTGAGCTGAATTTCACCATTCGGTTCGGTAAGCTTTAAACTATCAGCTATTGATTTACATAGCTGACTGGCAGGTGTCAAATCGTCCCAAATTGTTTGAGTCGGTGATAGACTACTACGCCGGGTCAACGTCGTCGGAGCTAGACGTGCCGTCACGACTTAAGTCCGTTGAGATCCGTTGCGCTTTTAATAAGTATCTTGAATGAAGGCCGAGTGTT
**************************************************.((((((..((..............(((((((((........)))))))))...............))..))))))...((((..(((..((((....)))).)))..))))......((((.((((.((((....)))).))..))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060666 160_males_carcasses_total |
SRR060675 140x9_ovaries_total |
M061 embryo |
SRR1106727 larvae |
M028 head |
SRR060667 160_females_carcasses_total |
V053 head |
V047 embryo |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060689 160x9_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................TGGAGATCAGGTGCGCTTTTAA............................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CGTTGAGATCCGTTGCGCTTTTA............................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AGCGTCGACTGCCTCCCTA...................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................GGTGCCCTCACCACTTAAG...................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CCGTAGCGCTATGAATAAGT....................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AGATTGTTTGAGTCGATGATC............................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CCGTAAGCTTCAAACTATCC............................................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AGATTGGTTGACTCGGTGAT............................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CGTAGCGCTATGAATAAGT....................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................AGCTTTAAACTAGCAGCC......................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACATAGCTGTCCCGCAGGT.............................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................TAAACTGTCAGCTAATGA.................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................ACCTTTCGGTTAGGTAAGTT....................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................TTGACTCGGTGATTGACT........................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:8217301-8217551 - | dvi_5698 | AACATCAGTGAACACAGCTCGCAGCTGACGGAGGCACTCGACTTAAAGTGGTAAGCCAAGCCATTCGAAATT---TGATAGTCGATAACTAAATGTATCGACTGACCGTCC--ACAGTTTAGCAGGGTTTAACAAA-CTCAGCCAC-TATCTGATGATGCGGCCCAGTTGCAGCAGCCTCGATCTGCACGGCAGTGCTGAATTCAGGCAACTCTAGGCAACGCGAAAATTATTCATAGAACTTACTTCCGGCTCACAA |
| droMoj3 | scaffold_6496:19024259-19024506 + | AGCATCAGTGAACATAGCTCGCAGCTAACTGAGGCACTCGACTTGAAGTGGTGAGCT-AGCTGTTCGATATATCCTGATAATAGCTT-T------TAATTATTATGAATTATTACAGTTTGGCAGGCTTCAATAGG-CTAAGCCAT-TTCCTAATGATGCGGCCCAGTTGCAGCAGTCTCGATTTGCATGGCACCGCCGAATTCAGGCAACTCTAGGCCCAGCAAAATTGATTCATAGAGCTGGCAAACGGTTCGCAA | |
| droGri2 | scaffold_15245:1677673-1677877 - | AGTACCACTGCACACAGCTCGCAGCTAACGGAGGCACTCGATTTGAAGTGGTAAATG-GATATATATACATATAC--ATATATATTT-T------TATTGACCATCTATCA--GCAGCTTAGCAGGCTGGTGAAAAGTTC-ACCATTTATGGATTGGATCGACCCAGCTGCAGCAGCCTCGATCTGGACGGCAGCTCGAAGATGAGGCAACTTTAAGC---------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:34 PM