
ID:dvi_5684 |
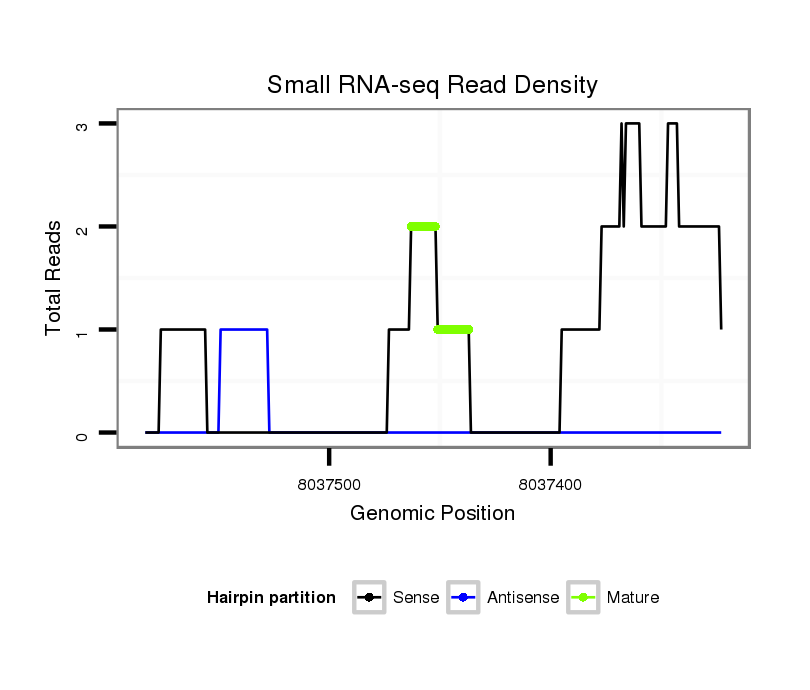
Coordinate:scaffold_12875:8037373-8037533 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
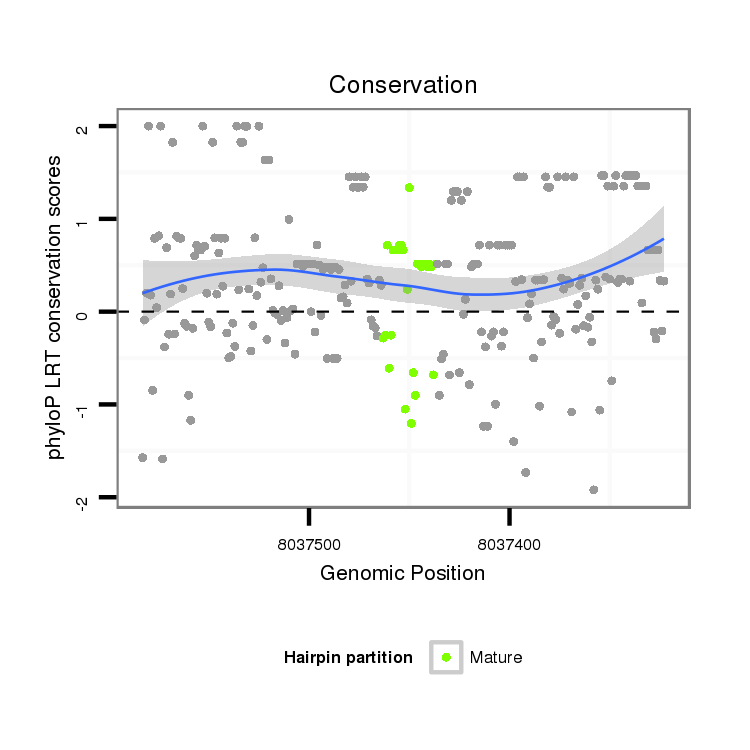
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
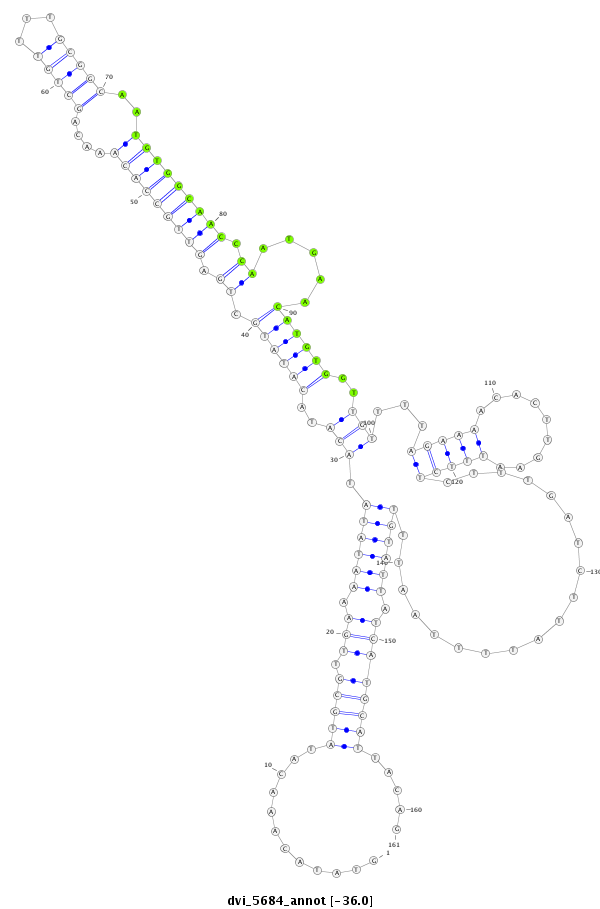
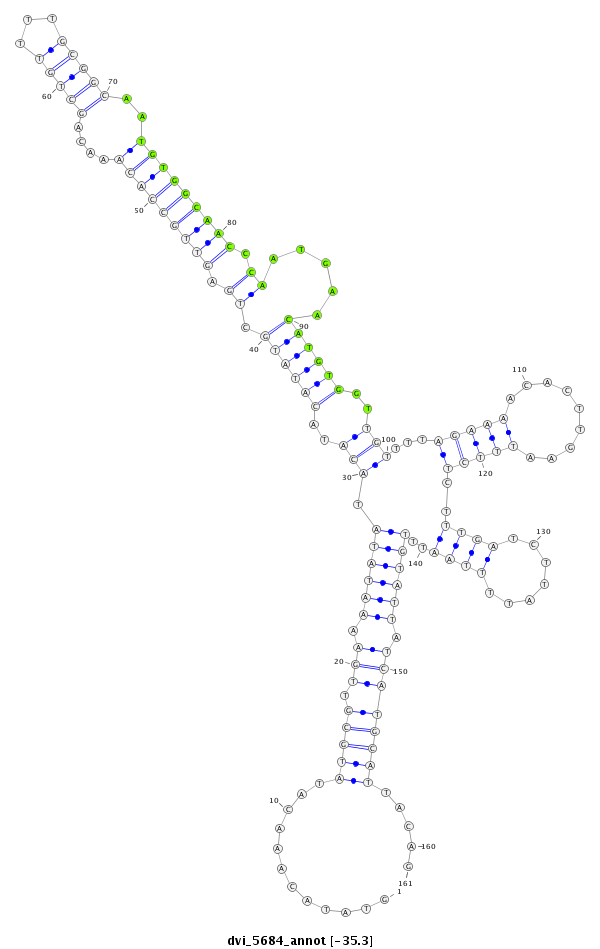
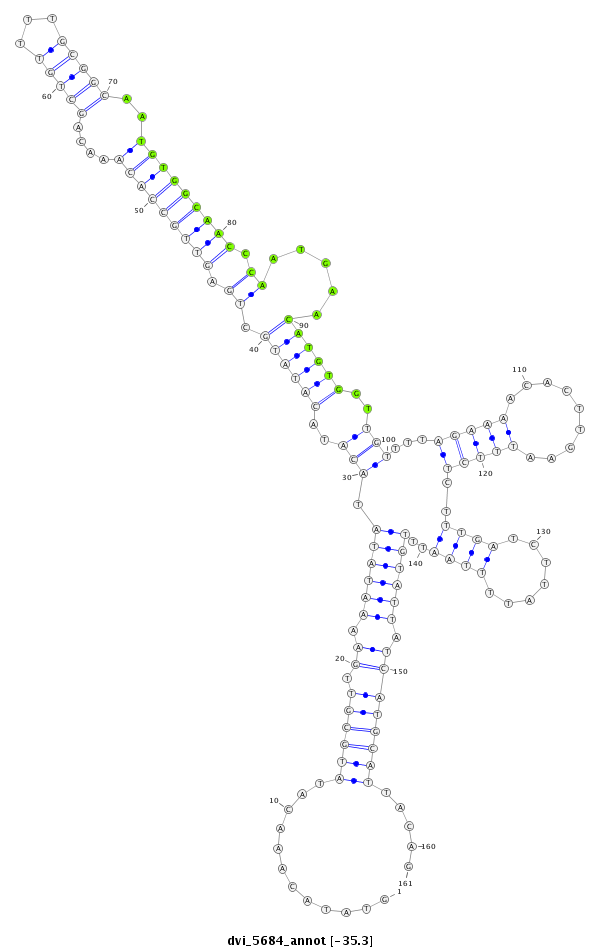
| -36.0 | -35.3 | -35.3 |
 |
 |
 |
exon [dvir_GLEANR_6124:1]; CDS [Dvir\GJ20706-cds]; CDS [Dvir\GJ20706-cds]; exon [dvir_GLEANR_6124:2]; intron [Dvir\GJ20706-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTGTGATATTTAGACCTAAAGTGCAAGTGCAGCCACAGCAAACTGTCAAGGTATACAAACATATGCGTTGAAAATATATACATACATATGCTGAGTTGCCACAAACAGCTGTTTTGCGGCAATGTGGCAACCCAATGAACATGTGGTTGTTTTAGAAAACACTTGAATTTCTCTTTGATCTTATTTTAATTTGTATTATCATGCATTACAGAAGGTAGCAGGCACTACTATTGCTCTCGGCAAAATACCTGCCGGAGCTAT **************************************************............((((((.((.((((((.(((..((((((.((.(((((((((....(((((...)))))..))))))))).)).....))))))..)))...(((((.........)))))...................)))))).)))))))).....************************************************** |
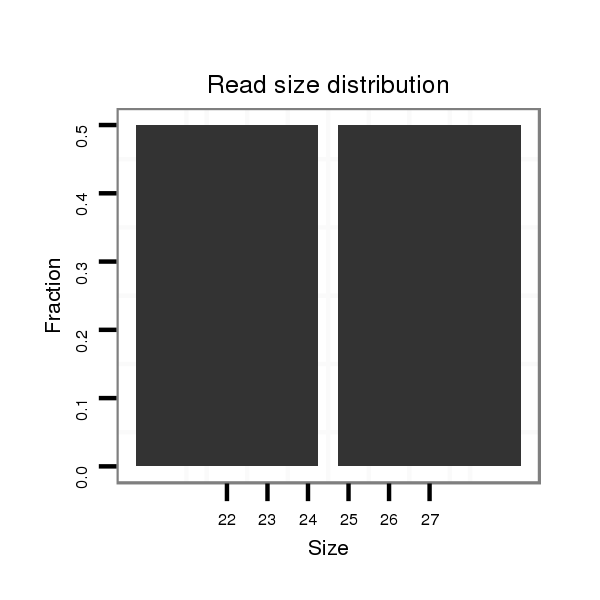
Read size | # Mismatch | Hit Count | Total Norm | Total | M061 embryo |
M047 female body |
GSM1528803 follicle cells |
V116 male body |
SRR060681 Argx9_testes_total |
M027 male body |
SRR060669 160x9_females_carcasses_total |
SRR060659 Argentina_testes_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060667 160_females_carcasses_total |
SRR060688 160_ovaries_total |
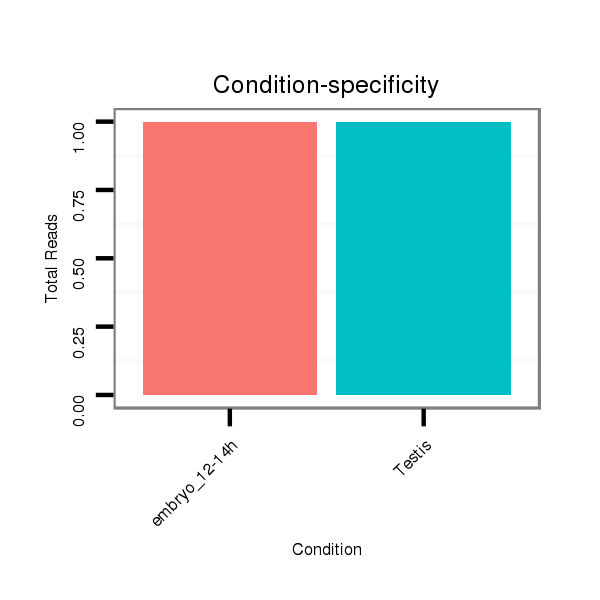
SRR1106723 embryo_12-14h |
V053 head |
SRR060685 9xArg_0-2h_embryos_total |
SRR060679 140x9_testes_total |
V047 embryo |
SRR060666 160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................CATAAAGTAGCAAGCACTACT................................ | 21 | 3 | 3 | 15.00 | 45 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAGGTAGAAAGCACTACT................................ | 21 | 3 | 2 | 8.00 | 16 | 12 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAAGTAGCACGCACTACT................................ | 21 | 3 | 2 | 3.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAAGTAGAAGGCACTACT................................ | 21 | 3 | 6 | 1.67 | 10 | 3 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TTACGGAAGGTAGAAGGCTC.................................... | 20 | 3 | 3 | 1.33 | 4 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................AATGTGGCAACCCAATGAACATGTGGT.................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......ATTTAGACCTAAAGTGCAAGT......................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAAGTAGCAGGCACTACT................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................TACAGAAGGTAGCAGGCA..................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................AAAATACCTGCCGGAGCTA. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ATTACGGAAGGTAGAAGGC...................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GCAGGCACTACTAATGCTCTCGGC.................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TAGCAGGCACTACTATTGCTCTCGGC.................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GTTTTGCGGCAATGTGGCAACC................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................ATTTGTATTATCATGCATTACAGAAGGT............................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................TCGGCAAAATACCTGCCGGAGCTAT | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ATTACGGAAGGTAGAAGGCTC.................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CAGAAAGTAGAAAGCACTACT................................ | 21 | 3 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GCAGGCACTACTATTGCTCTCGGC.................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AGAAAGTAGAAAGCACTACT................................ | 20 | 3 | 17 | 0.59 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAGGTAGAAAGCACTAC................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TAGAAAACTCATGATTTTCTCT....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CATAAAGTAGCAAGCACTAC................................. | 20 | 3 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................CCACACACCGCTGTTTTGAG............................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CACAAAGTAGCAAGCACTACT................................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CAGCCAAAGCAAACCGTCGAG................................................................................................................................................................................................................... | 21 | 3 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................ATAAGGTAGAAAGCACTACT................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................GTGCAGCCAGAGGAAACT......................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................TACGGAAGGTAGAAGGCT..................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................TTACGGAAGGTAGAAGGCT..................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGCATCCAAACATATGCGTC................................................................................................................................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GACACTATAAATCTGGATTTCACGTTCACGTCGGTGTCGTTTGACAGTTCCATATGTTTGTATACGCAACTTTTATATATGTATGTATACGACTCAACGGTGTTTGTCGACAAAACGCCGTTACACCGTTGGGTTACTTGTACACCAACAAAATCTTTTGTGAACTTAAAGAGAAACTAGAATAAAATTAAACATAATAGTACGTAATGTCTTCCATCGTCCGTGATGATAACGAGAGCCGTTTTATGGACGGCCTCGATA
**************************************************............((((((.((.((((((.(((..((((((.((.(((((((((....(((((...)))))..))))))))).)).....))))))..)))...(((((.........)))))...................)))))).)))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060665 9_females_carcasses_total |
SRR1106730 embryo_16-30h |
SRR060654 160x9_ovaries_total |
SRR060667 160_females_carcasses_total |
V116 male body |
V053 head |
M047 female body |
SRR060671 9x160_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............GATACCACGTTCACGTCTGTGTCGT............................................................................................................................................................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CGTCCGGGATGATAACGAGAG....................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TGTCGTTTGACAGTTCCATATG............................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TTTTGTGAACTTAAAGAGAAC..................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TGCCGTTACACTGTTGGG................................................................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CGTAAAGAGAAACTAGTATTAA........................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................AGTCTGTATACGCAAGTTTTA.......................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................AAGTTAAAGAGAAACTAGAAG.............................................................................. | 21 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................TTAAGGCGAAACTAGAATAG............................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................CACGGCGGGGTCGTTTGAG........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:8037323-8037583 - | dvi_5684 | CTGTGATATTTAGACCTAAAGTGCAAGTGCAGCCACAGCAAACTGTCAAGGTATAC-AAA--CATATGCGTTGAAAATATATACATACATAT---------GCTGA------GTTGCCA---------------------------------------------------CAAACAGCTGTTTTGCGGCAATGTGGCAACC-------------------------------------------------CAATG--AACATGTGGTTGTTTTA----------------------------------------------------------------GAAAACACTT--GAATTTCTCTTTGATCTTA---TTTTAATTTGTATT--ATCATGCATTACAGAAGGTAGCAGGCACTACTATTGCTCTCGGCAAAATACCTGCCGGAGCTAT |
| droMoj3 | scaffold_6496:19231487-19231744 + | CCATGATATTTAGACCTAAAATTGAAGTGCAGCAACAGCAAGCTGTCAAGGTATAGCACA-C-ATATCTA-------------------TAT---------ACTAACTTAATGCTACCA---------------------------------------------------CAAACAGCTGTTTCGAGGCGGTCCGGCAACG-------------------------------------------------CACTGGG-----------------CGTTCTGATTACATTTAAGGAATATTAGAATACAAT--------------------------------------------TTGTGTATTATTTTA---TTGAAACTGAAACTGAACAATGCATTACAGAAGATAGCAGGCGGCAGCATTGCGCTCGGCAAAATACCTGCCGGAGCTAT | |
| droGri2 | scaffold_15245:1527119-1527422 - | GTGTGGTATTCCGGTCTAAAGGGCCACTGCAGCCACAACAAATGTTAAAGGTATAC-AAA--CATTTGCAGACAGAATACATACATA-AAATG------------------------CA---------------------------------------------------TAAACAGCTGTTTTATGGCGATGTGGCAACCATTTGTGTATAAAAACGCAAATCAGCTGCTCGAAAATAATCTCGTAGTCCATAC--AACATGTGTTTCACTTC----------------------------------------------------------------GAAATCATTGACGAATTATGTTTAAATTATATTTTTTAAATTAA--TTG-AATATGCATAACAGAAGCTAACAGGCACAAGCATTGCGCTCGGCAAGATACCAGCCGGTTCTCC | |
| droBip1 | scf7180000396759:2810656-2810713 - | TCGTACCATTTCGGCCAAAGACAGAAG---ATTTCCAGCGAATACCAAAGGTAGGC-AAA--CA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302476:2900658-2900713 + | GCGTGGTTTTAAAACCAATAACCATTG---ATCCCCAGCAAATAATCAGGGTAAAC-TAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000491214:1009053-1009344 + | CCATGATTTTCCGCTCGAAAACATCGG---ATCCCCCGCAGATAACAAAGGTAAAT-AAAAT-GTATGCAAAA------------------AATACGAACT----------------------CCTATTTTTTGTTGTCATCCGGCGTTGCCAGATGGGCAAGAGGTGCACAAACAGCTGTTAAATAAC-----------A-------------------------------------------------------------------------TGTGCAGAGCAGAACAGCGAAATGTAA-ATCGCAATTGATAAAGTAATTTAATTTATTTACGCAGAAATCATTCGC------------------------GAAATTTGAACCGGGTAATGCCACAGTGAGGATAACTTCGGGTGGAATAGTACTCAACAAAATACCAGC------TAT | |
| droBia1 | scf7180000301754:5545904-5545982 - | CCGTGGTTATACGCCCGAAAACACAGG---ATCTTCAGCAAACTTTGAAGGTAATC-AGAATAGTCTGCAAAA------------------TGTATAAACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415345:366133-366212 - | CCGTGGTTTTACGCCCGAAAACACAGG---ATCCCCAGCAAATAATAAAGGTAAAATAGAACAGTCTGCAAAA------------------TGTATAAACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409672:1816942-1817249 + | ACGTTGTTTTTCGCCCGAAAACACTGG---ATCCGCAGCAAATAACAACGGTAAGC-AAA--AGTCTGCAAAATACAAG------------TATATAAACAA---------------CT---CCCCCTTTTTTGTTTTCATTCGGCGTTGCCAGATCGACTTGAGCTGCGCGAACAGCTGTCTAGTTAC-----------T-------------------------------------------------TATGGGA-----------------AGTGCCGAGCACAACAAATAAATGTAA-ATTTCAATTATTTAAGTAATTACTTTTCTTGGCACAGAAACCGTTTCC------------------------TAAATTAGAACCTGGAAATGCCACAATCAGGGTCACTTCAGGGGGCATAGTTCTCAACAAAATACCAGC------TGT | |
| droSim2 | 2r:13946468-13946766 - | dsi_18690 | CAGTGATCTTGAGCCCCGAAATACTAG---ATTCCCAAGATAACACAAAGGTAATC-AAA--AGTCTGTCAAA------------------TGTATAAACTA---------------TATTCTCATCCTTTTTGTTGTAATTCGGCGTTGCCAAATCGTCAAGAGATGCTCGACCAGCTGTTTAGTGAA-----------A-------------------------------------------------TACATGA-----------------TGTGCTGAACACAATTTTGAATTATAA-ATCATAATCAT------ACTTGTACTTATTTGCCCAGAAACCGTTTAC------------------------AAAATTGGACTCGGGAAGTGCATCAGTGAGAGTTAATGCAGGCGGTATAGTTCTTAACAAAATACCAGC------GAT |
| droSec2 | scaffold_1:10795557-10795583 - | T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAGTTCTTAACAAAATACCAGC------GAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/16/2015 at 08:28 PM