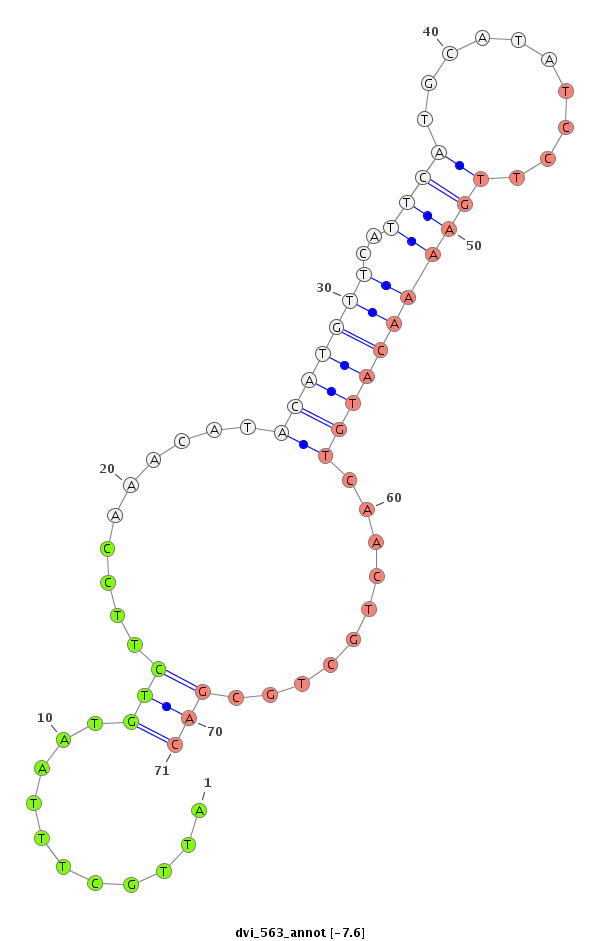
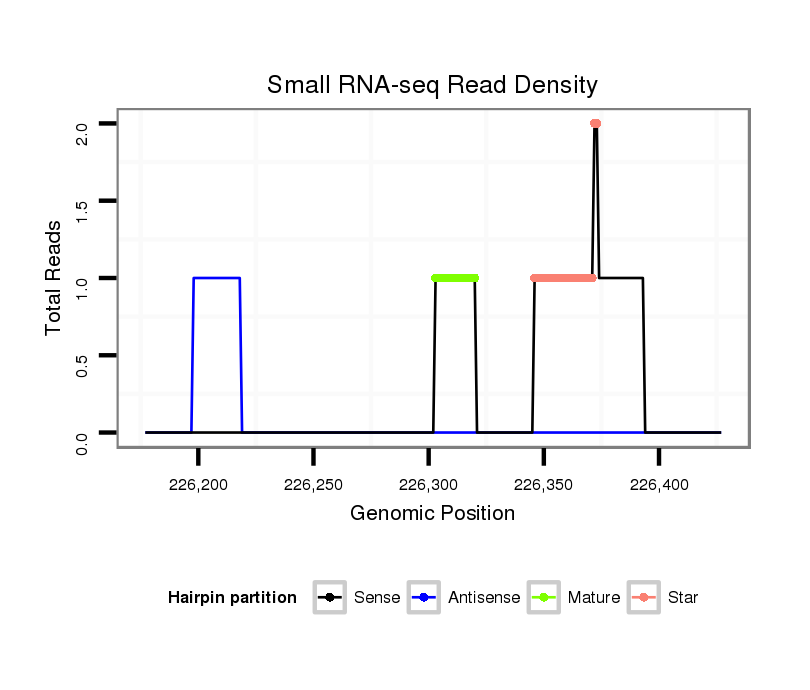
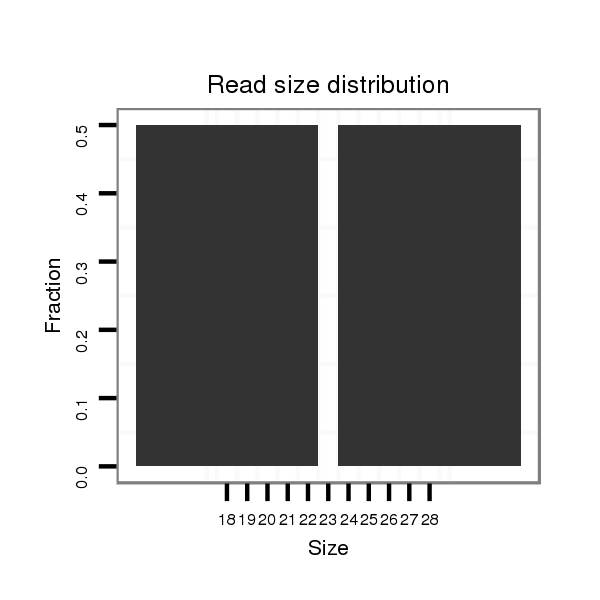
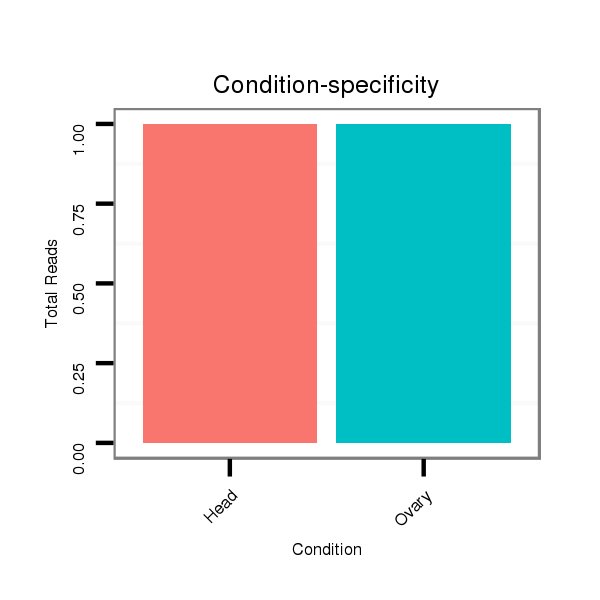
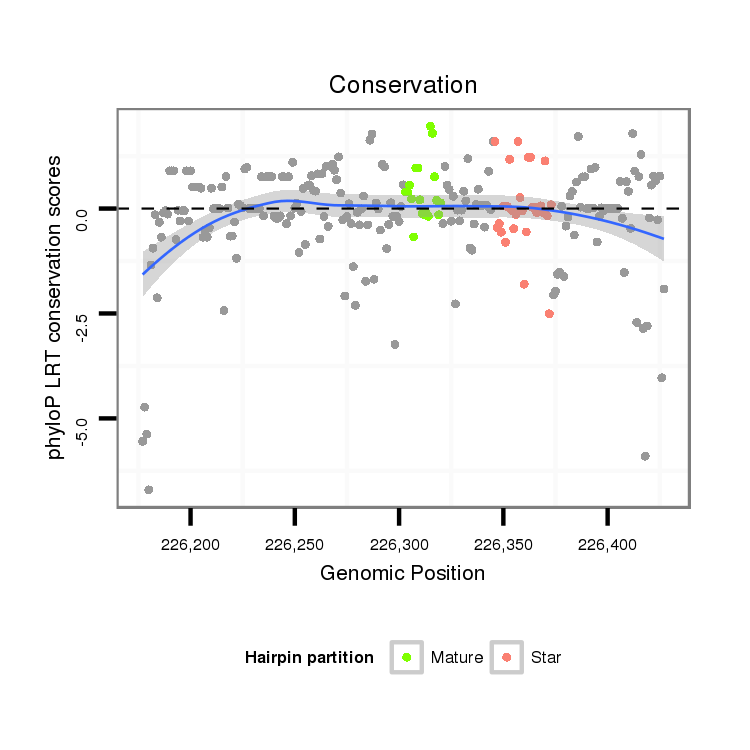
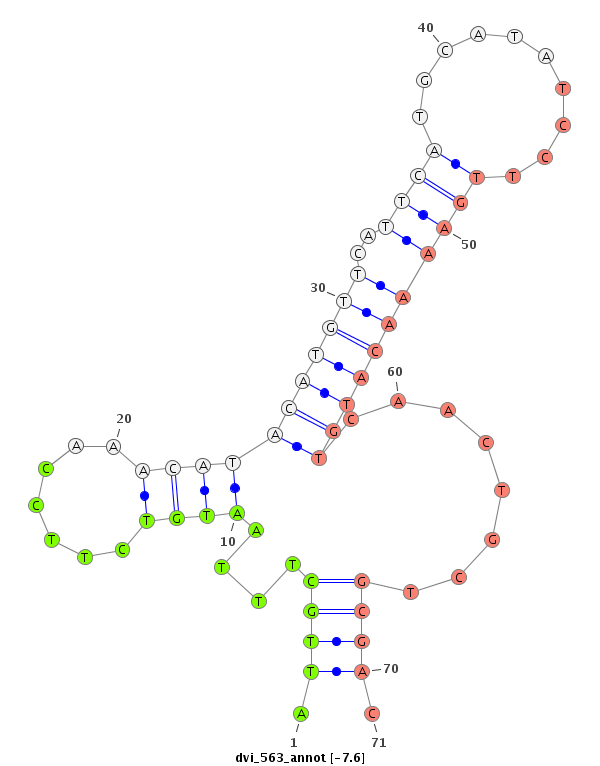
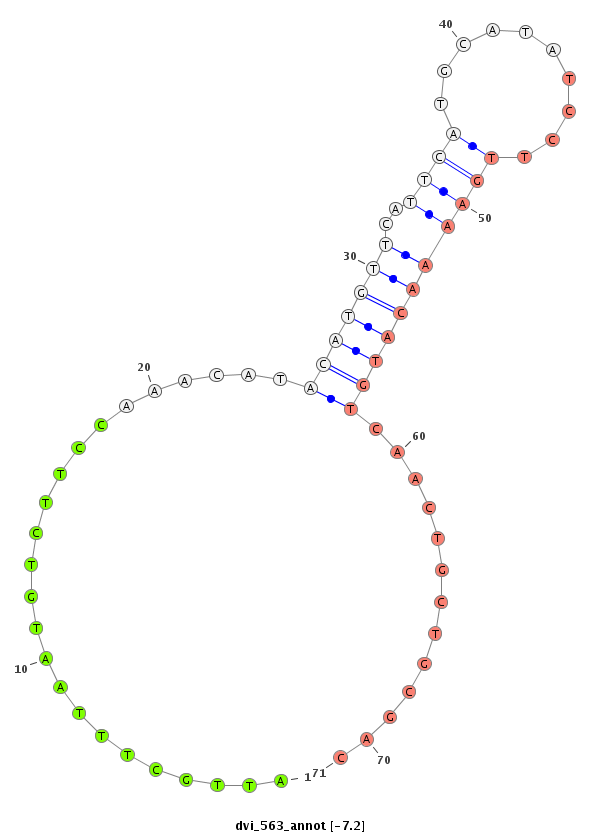
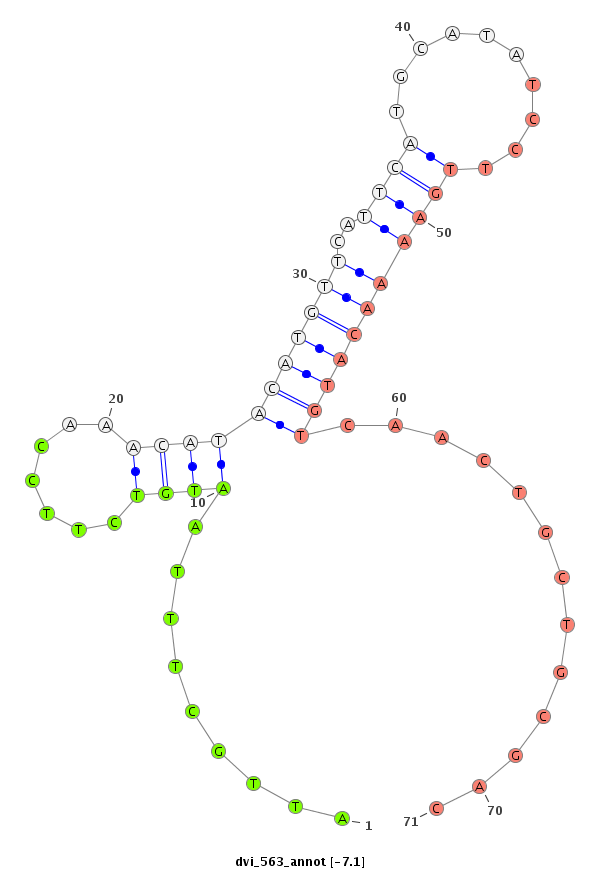
| CAGA---------------CACAC----------------------------------------------------------------------------------------------------------------A----------CA----CA---CAC-----A----CAC--ACAC--A------CAC--AAC----------------------ATCCCGCC---TCACCTCAC-------------------------------------------CGGCT---CTTTCTGTT-----------------------------------------TACTTTCTTTTCTGTATAC-TGTATATA--ATGCTCTGTA----C--------------AAAATCAAATCAACC----AA-------------------------------AT--------C-------AACCCCTCC-----------------------------------------------------------------------GCCCGCCCGTCCAAATGTAGAGCTTCAGTTCGAAGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------G--------GC------------------CATGTT--CGCATA-- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ...................................................................................................................................................................................................................................................................................................................................................................ATAC-TGTATATA--ATGCTCTGT.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 5.00 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCCAAATGTAGAGCTTCAGTT...................................................................................................................................................................................................................... | 21 | 1 | 4.00 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AATGTAGAGCTTCAGTTCGAAGGCGG............................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..................................................................................................................................................................................AC--A------CAC--AAC----------------------ATCCCGCC---TCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................TAC-TGTATATA--ATGCTCTGTA----......................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................................................................................TT-----------------------------------------TACTTTCTTTTCTGTATAC-................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CGTCCAAATGTAGAGCTTCAG........................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................CC----AA-------------------------------AT--------C-------AACCCCTCC-----------------------------------------------------------------------GCCCG............................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................CTGTATAC-TGTATATA--ATGC................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGTAGAGCTTCAGTTCGAAGGCGGC............................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................A-------------------------------AT--------C-------AACCCCTCC-----------------------------------------------------------------------GCCCGCCC............................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CGCC---TCACCTCAC-------------------------------------------CGGCT---CTT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................AAATCAACC----AA-------------------------------AT--------C-------AACCCCT............................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................T-----------------------------------------TACTTTCTTTTCTGTATAC-T................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CGAAGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------G--------GC------------------................ | 20 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................................................................................ACTTTCTTTTCTGTATAC-TG............................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................CAAATCAACC----AA-------------------------------AT--------C-------AACCCC.............................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................C-------AACCCCTCC-----------------------------------------------------------------------GCCCGCCCGTC......................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................TCTGTATAC-TGTATATA--ATGC................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AATGTAGAGCTTCAGTTCGAA.................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CGCCCGTCCAAATGTAGAGCT............................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTAGAGCTTCAGTTCGAAGGC............................................................................................................................................................................................................... | 21 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AAC----------------------ATCCCGCC---TCACCTCAC-------------------------------------------C.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCTTCAGTTCGAAGGCGGCG........................................................................................................................................................................................................... | 21 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................TATATA--ATGCTCTGTA----C--------------AAAA...................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................AC--ACAC--A------CAC--AAC----------------------ATCCCGCC---................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CAC--AAC----------------------ATCCCGCC---TCACCTC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CAC--ACAC--A------CAC--AAC----------------------ATCCCGC....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................CTCAC-------------------------------------------CGGCT---CTTTCTGTT-----------------------------------------TA................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................T-----------------------------------------TACTTTCTTTTCTGTATAC-C................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TCCCGCC---TCACCTCAC-------------------------------------------CGGC........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................AACC----AA-------------------------------AT--------C-------AACCCCTCC-----------------------------------------------------------------------GCCGA............................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................TATAC-TGTATATA--ATGCTCT................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................T--------C-------AACCCCTCC-----------------------------------------------------------------------GCCCGCCCGT.......................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................AT--------C-------AACCCCTCC-----------------------------------------------------------------------GCCCGC.............................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGTTCGAAGGCGGCGCTGCA...................................................................................................................................................................................................... | 20 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................TATAC-TGTATATA--ATGCTCTG............................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................TGTATAC-TGTATATA--ATGCTC................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................TCCCGCC---TCACCTCAC-------------------------------------------CGGCT---....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................CAACC----AA-------------------------------AT--------C-------AACCCCTCC-----------------------------------------------------------------------GC.................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................CTTTCTTTTCTGTATAC-TGTA............................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTCAGTTCGAAGGCGGCGCTT........................................................................................................................................................................................................ | 21 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GAAGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------G--------G................................... | 18 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------G--------GC------------------A............... | 18 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTTCAGTTCGAAGGCGGCGC.......................................................................................................................................................................................................... | 20 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTCAGTTCGAAGGCGGCGCT......................................................................................................................................................................................................... | 20 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGTTCGAAGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------............................................. | 21 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTAGAGCTTCAGTTCGAAGG................................................................................................................................................................................................................ | 20 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCTTCAGTTCGAAGGCGG............................................................................................................................................................................................................. | 19 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCGAAGGCGGCGCTGCAC--------------------------------------------------------------------------------------------------------------------------------------------------------G--------GC------------------................ | 21 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TAGAGCTTCAGTTCGAAGGCGGCG........................................................................................................................................................................................................... | 24 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................C-----A----CAC--ACAC--A------CAC--AAC----------------------AT............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAATGTAGAGCTTCAGTTTCC................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |