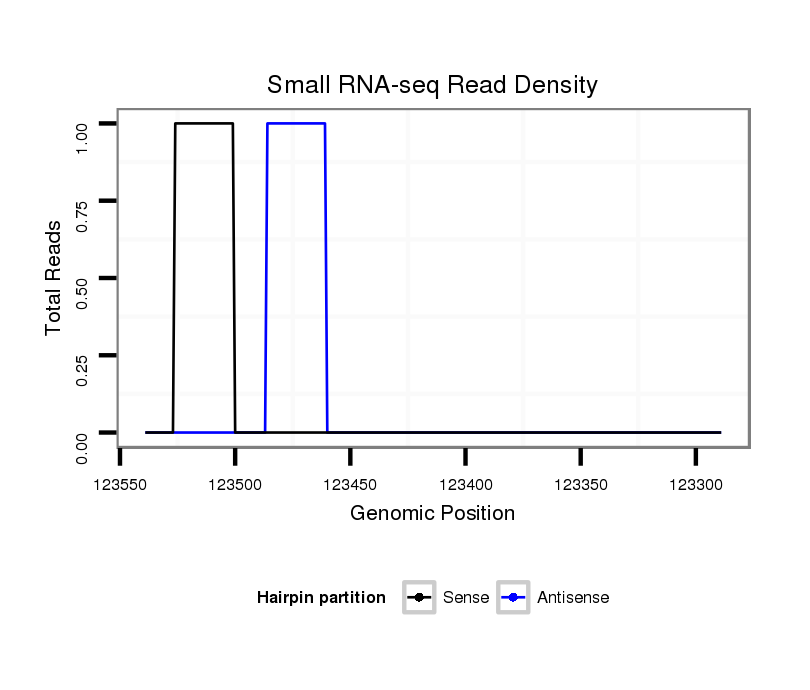
ID:dvi_558 |
Coordinate:scaffold_10322:123339-123489 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dvir\GJ16058-cds]; exon [dvir_GLEANR_16478:2]; intron [Dvir\GJ16058-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GCGCGTTCTGGAACCGCTTCTACACGTCTAGCAATGAGGAAAACGATTCCGTAAGTTATTCCAACAATGGATCAGTTTAATATGCGCATACTTTAAATGCGTATGAAGGATATTAGGTTTTTGTGTGTATATAGCAGGCAGAAGGGAGCCTGTCTTGTTCAGGACATTAGAGCTGAGTCGATATGGTCATGTAGCCATGAATACCTTAAAAGTAGGCTCTCGCAATATGAAAAGTATCGCAAAAAGAACAA **************************************************.((((((..((((....))))..)))))).(((((((((.......)))))))))...(((.((((.(((.(((.((.(((..((((((........))))))..))).))..)))..))).)))))))..((((((......))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060682 9x140_0-2h_embryos_total |
V116 male body |
SRR060689 160x9_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
M027 male body |
SRR060663 160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
M047 female body |
SRR060685 9xArg_0-2h_embryos_total |
V053 head |
SRR060680 9xArg_testes_total |
SRR060681 Argx9_testes_total |
SRR060667 160_females_carcasses_total |
GSM1528803 follicle cells |
SRR060679 140x9_testes_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
SRR1106729 mixed whole adult body |
SRR060671 9x160_males_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR1106727 larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................GGCAGAAGGGAGCTTGACT................................................................................................ | 19 | 2 | 2 | 6.50 | 13 | 10 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGGCACAAGGGAGCTTGACT................................................................................................ | 20 | 3 | 8 | 3.75 | 30 | 6 | 10 | 0 | 0 | 2 | 1 | 1 | 1 | 3 | 2 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGGCACAAGGGAGCTTGAC................................................................................................. | 19 | 3 | 20 | 2.00 | 40 | 0 | 38 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................GGCAGAAGAGAGCTTGACT................................................................................................ | 19 | 3 | 20 | 1.75 | 35 | 6 | 7 | 1 | 0 | 4 | 0 | 7 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| .............CCGCTTCTACACGTCTAGCAATGAGG.................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GGCACAAGGGAGCTTGTCT................................................................................................ | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGGCAGAAGAGAGCTTGACT................................................................................................ | 20 | 3 | 12 | 0.92 | 11 | 0 | 0 | 0 | 1 | 1 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................GGCAATGAGGAGAACGATT........................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................GATGAAAACGATTCTGTA...................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GGAAAAGGTTTCCGTAGGT................................................................................................................................................................................................... | 19 | 3 | 17 | 0.12 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................GTACCCCTGAAGACCTTAA.......................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GGCAGAAGGGAGCTTGAC................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................ATTGCCACAATGGATCAGAT.............................................................................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGGCAGAAGGGAGCTTGAC................................................................................................. | 19 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GGTAGAAGGGAGCTTGACT................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGGCATAAGGGAGCTTGAC................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GCACAAGGGAGCTTGTCTG............................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGCGCAAGACCTTGGCGAAGATGTGCAGATCGTTACTCCTTTTGCTAAGGCATTCAATAAGGTTGTTACCTAGTCAAATTATACGCGTATGAAATTTACGCATACTTCCTATAATCCAAAAACACACATATATCGTCCGTCTTCCCTCGGACAGAACAAGTCCTGTAATCTCGACTCAGCTATACCAGTACATCGGTACTTATGGAATTTTCATCCGAGAGCGTTATACTTTTCATAGCGTTTTTCTTGTT
**************************************************.((((((..((((....))))..)))))).(((((((((.......)))))))))...(((.((((.(((.(((.((.(((..((((((........))))))..))).))..)))..))).)))))))..((((((......))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060656 9x160_ovaries_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060680 9xArg_testes_total |
M047 female body |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................TCAATAAGGTTGTTACCTAGTCAAAT............................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................GTATCGGCCGTCTTCCCT........................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .....................................................TAAACAAGGTTGTTACTTAGT................................................................................................................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................CGTCCGTCTTCCCTACCAC................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_10322:123289-123539 - | dvi_558 | GCGCGTTCTGGAACCGCTTCTACACGTCTAGCAATGAGGAAAACGATTCCGTAAGTTATTCCAACAATGGATCAGTTTAATATGCGCATACTTTAAATGCGTATGAAGGATATTAGGTTTTTGTGTGTATATAGCAGGCAGAAGGGAGCCTGTCTTGTTCAGGACATTAGAGCTGAGTCGATATGGTCATGTAGCCATGAATACCTTAAAAGTAGGCTCTCGCAATATGAAAAGTATCGCAAAAAGAACAA |
| droMoj3 | scaffold_6680:2168232-2168289 - | TTGCCTTTTGGAATCGCTATTACACCTCCAGCAATGAGGAAAACGATTCCGTAAGTCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:385058-385119 + | ATGTGGCTTGGAATCGCGACTATTTCTCGAGCAATGCGGAAACCGAATCCGTAAGTTGATCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 07:47 PM