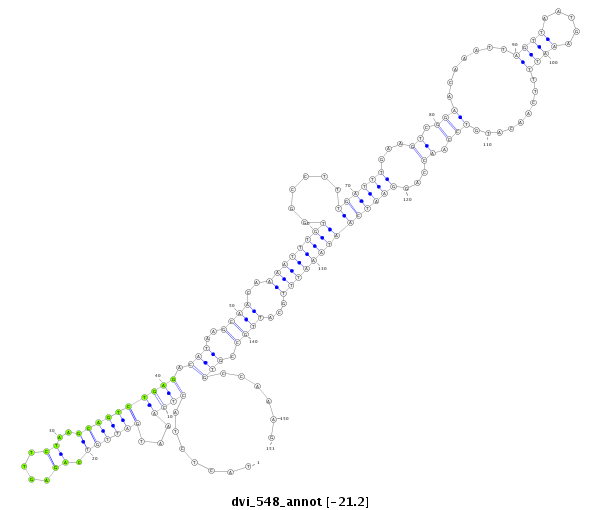
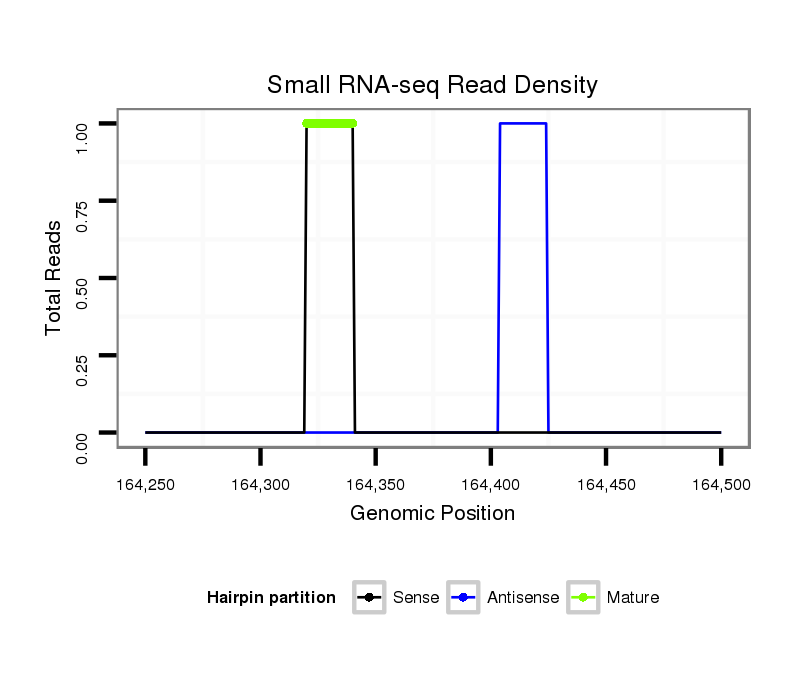
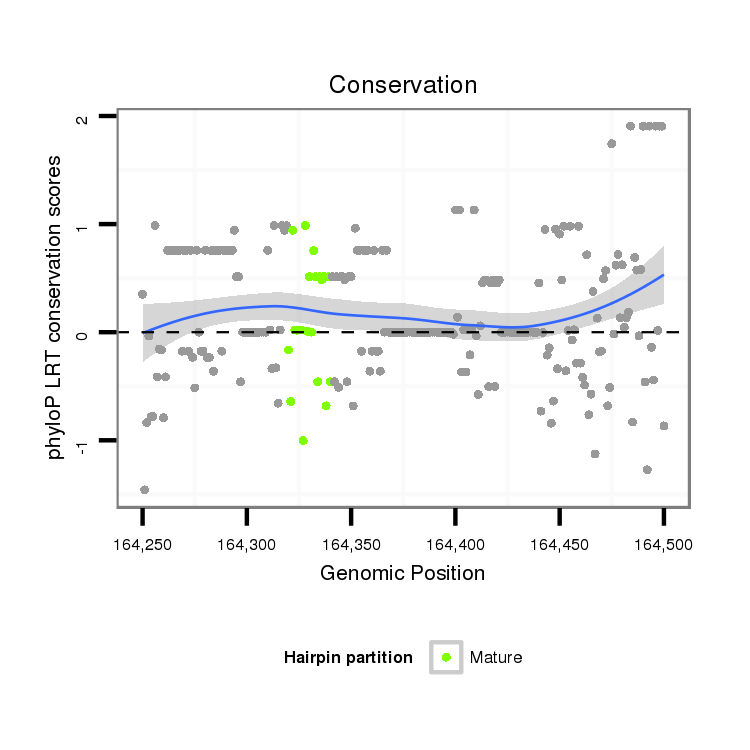
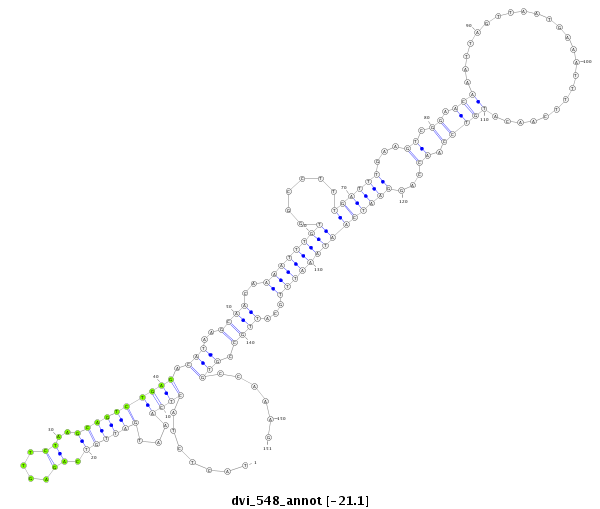
ID:dvi_548 |
Coordinate:scaffold_10322:164300-164450 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.1 | -21.1 | -21.0 |
 |
 |
 |
exon [dvir_GLEANR_16501:2]; CDS [Dvir\GJ16080-cds]; intron [Dvir\GJ16080-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGTTGCATTCCATATTATTCCTTTTGTTAGATACTATAACTTTTGTTAAATACTCTACTCAAATGATTGTCAGAGTTCTAAGCAGTCTGAGACATAAGCAACAAAATTTGTGGCCTTTGATTTGAAGTCGGAACAAATTAGTTAATGAAATTTTCAACATGTCCAACCAGGAATCAATAAATTTGCATTGCCGTGCCAAAGATACTGAAAAAATCGTCCAAGTAATCCATGGTATGTCCGAACTGGTCAAT **************************************************.......((((...((((((.((....))..)))))))))).(((..((((..((((((((......((((((...((.(((.......((((.....)))).........))).))...))))))))))))))...)))).)))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
SRR060682 9x140_0-2h_embryos_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060679 140x9_testes_total |
M047 female body |
M061 embryo |
M028 head |
SRR060666 160_males_carcasses_total |
V053 head |
SRR060657 140_testes_total |
SRR060660 Argentina_ovaries_total |
SRR1106729 mixed whole adult body |
SRR060663 160_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
SRR060658 140_ovaries_total |
SRR060659 Argentina_testes_total |
SRR060687 9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060661 160x9_0-2h_embryos_total |
SRR060689 160x9_testes_total |
SRR060671 9x160_males_carcasses_total |
M027 male body |
SRR060676 9xArg_ovaries_total |
SRR060669 160x9_females_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................GAAGTCGGAACAAGGTAGC............................................................................................................. | 19 | 3 | 14 | 5.57 | 78 | 10 | 22 | 12 | 1 | 3 | 3 | 9 | 0 | 2 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 2 | 3 | 1 | 2 | 0 | 1 | 1 | 0 |
| ..........................................................................................................................TGAAGTCGGAACAAGGTAGC............................................................................................................. | 20 | 3 | 2 | 5.00 | 10 | 5 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TCTGCGACCTGAGCAACAAAA................................................................................................................................................. | 21 | 3 | 4 | 1.50 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GTGAAGTCGGAACAAGTTAGC............................................................................................................. | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GTGAAGTCGGAACAAGGTAG.............................................................................................................. | 20 | 3 | 3 | 1.00 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................GAAGTCGGAACAAGGTAG.............................................................................................................. | 18 | 2 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................CAGAGTTCTAAGCAGTCTGAG................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TGAAGTCGGAACAAGGTAG.............................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TGGGGTGAAGTCGGAACAA................................................................................................................... | 19 | 3 | 13 | 0.85 | 11 | 6 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........GATATTATTCCTTGTGTTTGAT........................................................................................................................................................................................................................... | 22 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TGAAGTCGTAACAAGGTAG.............................................................................................................. | 19 | 3 | 14 | 0.29 | 4 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GGGGTGAAGTCGGAACAA................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................ATTCCTTGTGTTTGATAC......................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................GAAGTCGAAACAAAGTAG.............................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TGCTGAAAAAATTGTCCAA.............................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................AAATGGTCCAGGTGATCCAT..................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................CATGTCGAACCAGGTATG............................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCAACGTAAGGTATAATAAGGAAAACAATCTATGATATTGAAAACAATTTATGAGATGAGTTTACTAACAGTCTCAAGATTCGTCAGACTCTGTATTCGTTGTTTTAAACACCGGAAACTAAACTTCAGCCTTGTTTAATCAATTACTTTAAAAGTTGTACAGGTTGGTCCTTAGTTATTTAAACGTAACGGCACGGTTTCTATGACTTTTTTAGCAGGTTCATTAGGTACCATACAGGCTTGACCAGTTA
**************************************************.......((((...((((((.((....))..)))))))))).(((..((((..((((((((......((((((...((.(((.......((((.....)))).........))).))...))))))))))))))...)))).)))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
M028 head |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060662 9x160_0-2h_embryos_total |
V047 embryo |
V053 head |
SRR060665 9_females_carcasses_total |
SRR1106729 mixed whole adult body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................GTTGTACAGGTTGGTCCTTAG............................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................ACCAAACAGGCTTGAGTAGTT. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................AAAACAATTTATGAGTAGAG............................................................................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AGACGCTGGACTCGTTGTTTT................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TCAGACGCTGGACTCGTTGTT................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CTCAAGGTTGGTAAGACTC................................................................................................................................................................ | 19 | 3 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GCAGGTTAATGAGGTAGCA.................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................TTCATTAGGTACCTAGCAG............. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................CTTTGATATTGAAAGCAA............................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................CTAAAGATTGGTAAGACTC................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TTAGCGGCACGGTTTC.................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_10322:164250-164500 + | dvi_548 | AGTTGCATTCCA----TATTATTCCTTTTGTTAGATACTATAACTTTTGTTAAATACTCTACTCAAATGATTG-----------TCAGAGTTCTAAGCAGTCTGAGACATAAGCAACAAAAT--------------TTGTGGCCTTTGATTTGAAGTCGGAACAAATTAGTTAATGAAATTTTCAACATGTCCAACCAGGAATCAATAAATTTGCATTGCCGTGCCAAAGATACTGAAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATCGTCCAAGTAATCCATGGTATGTCCGAACTGGTCAAT |
| droMoj3 | scaffold_6680:2213229-2213318 + | AACTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTCCATCATGTCCAATCAAGA------------------CAGTGTTCAGGATGCTGAGAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTGGTTCAAGTGATCTTTGGCATGGCCGAGTTGCTCAAC | |
| droGri2 | scaffold_15110:338218-338466 - | ACTTGAAACTGT------------------------------------GTTG-------------AGTATTTGTTATATTTGCTTTCGAGTTGTAA--AATCTTAAATAAAAGTAAAA------------------------------------------------------------CTTTCACTTGTCGT--------------------------------------------------------------------------CGACAC-----------------------TTGATTAATATATTACATACTTGATTTACAGTCGTTTTTTGTTTTGCTTTTTTTTTTAATTTCGAGTAATTTTGAAAAATGCCCAAACCATAAGATGC-AAGTGGAAAATATATAATATATAATCTGCGCAAAAACATTCACGGCATGTCGGAGTTGTTCAAT | |
| droAna3 | scaffold_13337:434277-434306 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGGTGCACGCCTTGTCCGACCTAGTGAAC | |
| droBip1 | scf7180000395155:325323-325411 - | AAGA-----------------------------------------------------------------------------------------------------------------A------------------------------------------------------------ATCTTCCTTATGCC--------------------------------------------------------------------------CGGGACAAA-------------------------------------------------------------------------------------------------AACACAGAAACAGTCGGATTTCAA-------------------------GCTGACCGTGCTGCACGCCCTATGCGATTTGTTGAAT | |
| droFic1 | scf7180000454105:715905-715928 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATGCCATTTCGGAAATAGTGAAT | |
| droRho1 | scf7180000780275:59203-59256 + | TTGTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCCGAAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------AGA-------------------------CTTAATAGTGCTGCATGCCATGTCGCAATTAGTGAAC | |
| droEug1 | scf7180000409466:116461-116490 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCTGCATGCCATCTCGGAATTGGTCAAT | |
| droYak3 | 3L:9546906-9546929 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATAATATATCGGAAATAGTGAAC | |
| droEre2 | scaffold_4784:2404473-2404689 + | AATAGCATCCCAGAAATATTATTTCTCTACT-GAAATCGATAGCTTTTG---------------AGATACCTG-----------TCAGGACCATT--C--------------------AAGTTTGACCTTAAACTATTTCGATAT-T------------------------------------------------------------------------------------TATTTGGAGTTCGTATCAAGCTGTCTCAAAATTCGTAGAATAAAGATGTCCAAAAAATCGACACTCGATGAAC----------------------CCGACGA------------------------------------------------------------TCG-------------------------CTTCACTGTCTTGCATAATATATCGGAATTAGTGAAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 07:48 PM