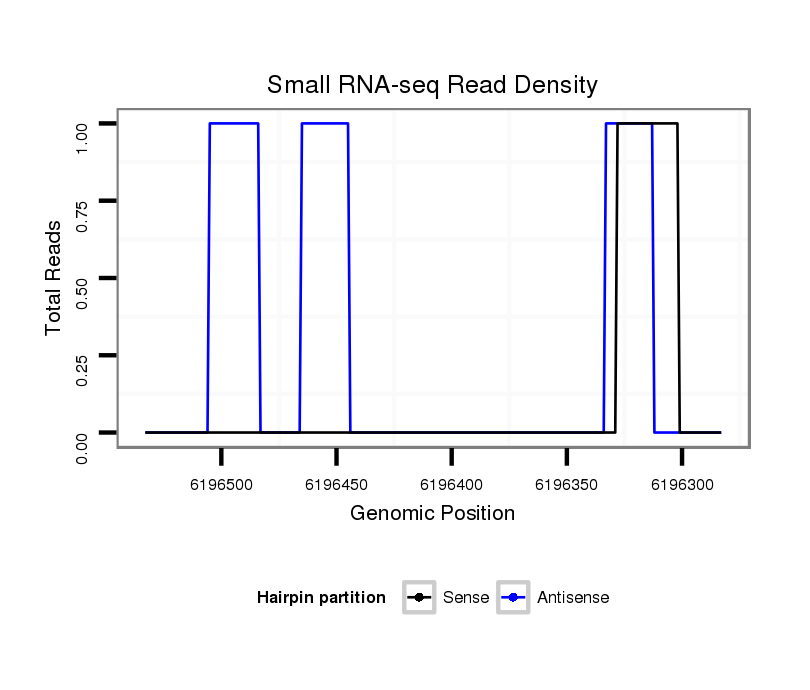
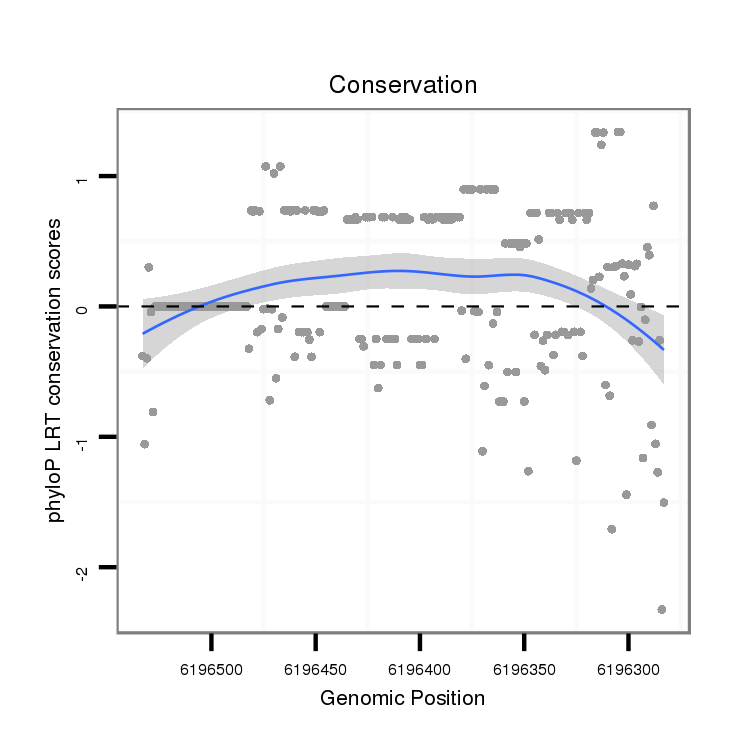
ID:dvi_5450 |
Coordinate:scaffold_12875:6196333-6196483 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_6226:3]; CDS [Dvir\GJ20811-cds]; intron [Dvir\GJ20811-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACAATTAGATTAGCCGGACAACCCTTTTGGGCGGACATTTTTAGTCAAATTTTGATGAATCATCCAAAATACATACCCATACAACGGACTCCTTTTAAGCGGACAGAAATAACCCAATTGTCTCTGTCCGTTTCAAAGAGTTTTCACTGTATATACATAGTTAGTTATTTATCCATTAACACAATGTTTTACTACAAGCAGCCACTGCATGAAGATCAAGGAGACCATGGACTATATAAACGAGGTGGTCG **************************************************...((((...))))..............(((((..((((((.(((.(((((((((((.((((.....)))).))))))))))).)))))))))....)))))..((((.((((((.........))))))...))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060680 9xArg_testes_total |
M047 female body |
M061 embryo |
M027 male body |
V047 embryo |
V116 male body |
SRR060671 9x160_males_carcasses_total |
GSM1528803 follicle cells |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TGCATGAAGATCAAGGAGACCATGGAC................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TTTAAGCGGACCGGAATATC.......................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................ATAGTTAGTTACGTATCCAAT.......................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................GATCCATTAACACAGTGGTT.............................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GTAGCCACTTCATGAAGA.................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GAAGATCAAGACGACAATGG..................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................CATACTACGGGGTCCTTT............................................................................................................................................................ | 18 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................ATCATCCCAAGTTCATACC.............................................................................................................................................................................. | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CGGAAAACCCTTTTGAG............................................................................................................................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................ATAGTTAGTTACGTAGCCA............................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TGTTAATCTAATCGGCCTGTTGGGAAAACCCGCCTGTAAAAATCAGTTTAAAACTACTTAGTAGGTTTTATGTATGGGTATGTTGCCTGAGGAAAATTCGCCTGTCTTTATTGGGTTAACAGAGACAGGCAAAGTTTCTCAAAAGTGACATATATGTATCAATCAATAAATAGGTAATTGTGTTACAAAATGATGTTCGTCGGTGACGTACTTCTAGTTCCTCTGGTACCTGATATATTTGCTCCACCAGC
**************************************************...((((...))))..............(((((..((((((.(((.(((((((((((.((((.....)))).))))))))))).)))))))))....)))))..((((.((((((.........))))))...))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
SRR060677 Argx9_ovaries_total |
SRR060679 140x9_testes_total |
V116 male body |
SRR060681 Argx9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR1106729 mixed whole adult body |
V053 head |
SRR060664 9_males_carcasses_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............CGGCCTGTTGGAAAAACCCGCCTGTA..................................................................................................................................................................................................................... | 26 | 1 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................CCTGAGGAAAATTCGCCTGTCA................................................................................................................................................ | 22 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGGCCTGTTGGAGAAACCCGCCTGTA..................................................................................................................................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CCCGCCTGGAAAAATCAGTTTA......................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CGGCCTGTTGGACAAACCCGCCTGTA..................................................................................................................................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TATGTATGGGTATGTTGCCTG.................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CAGCCTGTTGGAAAAACCCGCCTGTA..................................................................................................................................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CCCGCCTGTAAAAATCAGTTTA......................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CGGTGACGTACTTCTAGTTCC.............................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................ATTGGGTTAACAGAAACA............................................................................................................................ | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GGGTTACAAATTGATGCTCG.................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGTTATTGGGTTAACAGAAACA............................................................................................................................ | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TGGCTATGTTGCCGGCGGA.............................................................................................................................................................. | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........TCGGCCTGTTGCGAA................................................................................................................................................................................................................................. | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................GTCTCTATTGCGTTGACAGA................................................................................................................................ | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................AGTTCAATAGTACTTAGTAG........................................................................................................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................CCTGTGTTTATTGGGTAAAAA.................................................................................................................................. | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:6196283-6196533 - | dvi_5450 | ACAATTAGATTAGCCGGACAACCCTTTTGGGCGGACATTTTTAGTCAAATTTTGATGAATCATCCAAAATACATACCCATACAACGGACTCCTTTTAAGCGGACAGAAATAACCCAATTGTCTCTGTCCGTTTCAAAGAGTTTTCACTGTATATA----------CATAG--TTAGTTATTTATCCATTAACACAATGT----------TTTACTACAAGCAGCCACTGCATGAAGATCAAGGAGACCATGGACTATATAAACGAGGTGGTC---G |
| droMoj3 | scaffold_6496:23839365-23839480 + | TATATA---------------------------------------------------------------------------------------------------------------------------------------------------TA----------TATAGATTTACGT-TTTAGAAACTAATACACTCTTCTTTTTTTTTTT--ACTAAATAGCCATTGCCTGAAGATCAAAGACACTATGGACTATATAGATGAGGTAGTTAACG | |
| droGri2 | scaffold_15112:1085770-1085845 - | ACAAAC---------------------------------------------C-------------------------------------------------------------------------------------------------------------------------------------------------TT----------TATATTCCAAACAGTTACTGTGTACAGATTAAGGAGAAAATGGACATTTTAAATGAGTTAATGAAAA | |
| droWil2 | scf2_1100000004762:4847755-4847864 + | AAAATA---------------------------------------------------CGTTCCCACAG------------------------------GCGGACGATAATACTGAAACCATTGCTGTCCACCCACAGGAGCTTTCACTGTATACAGCATGAGCGATATAA--CAATTTCTGTT-----------------------------------------------------------------------------------TCAATTAACA | |
| droPer2 | scaffold_4:6384901-6384906 - | ACAATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13417:820098-820107 + | GAGG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGGCC---A | |
| droBip1 | scf7180000395863:7753-7807 - | CAAAAT---------------------------------------------TTGACGCGTCGCCTCAAATACAGATTTACTAAACAGA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGCTTTTAAG---C | |
| droEle1 | scf7180000490214:1592303-1592335 - | AGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGATGAGGAGGATGAAAAGAAGTTGGAG---G | |
| droEug1 | scf7180000409095:116081-116121 + | GGAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAAGGAGAAAATGGCCAACATAACCGAGATTATT---G | |
| droEre2 | scaffold_4784:8479663-8479702 - | AAGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAGGACACCATGGACCACATCCATCAGGTGCAG---C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 04:25 AM