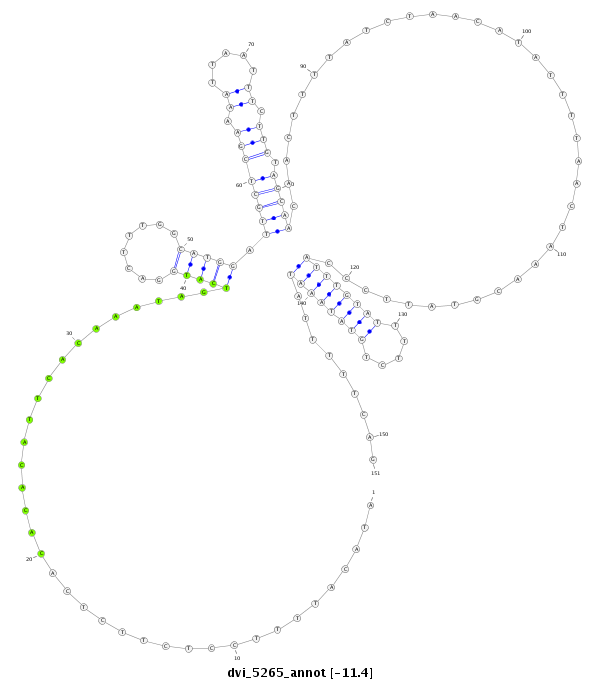
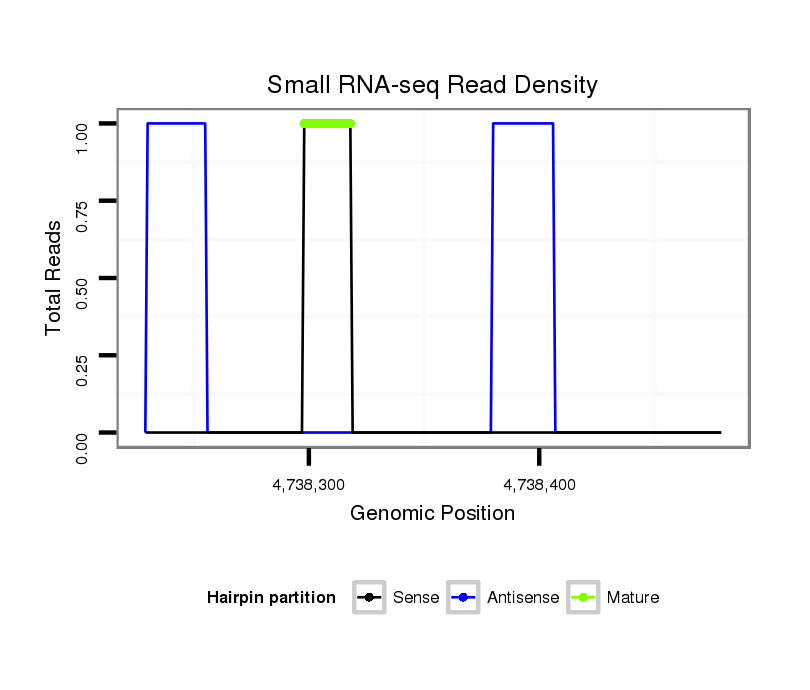

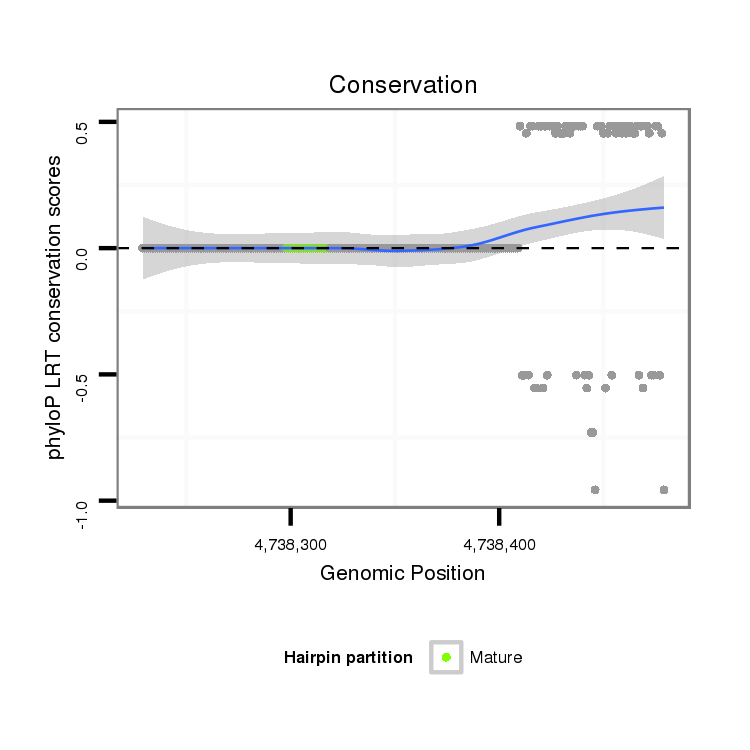
ID:dvi_5265 |
Coordinate:scaffold_12875:4738279-4738429 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -11.1 | -10.9 | -10.9 |
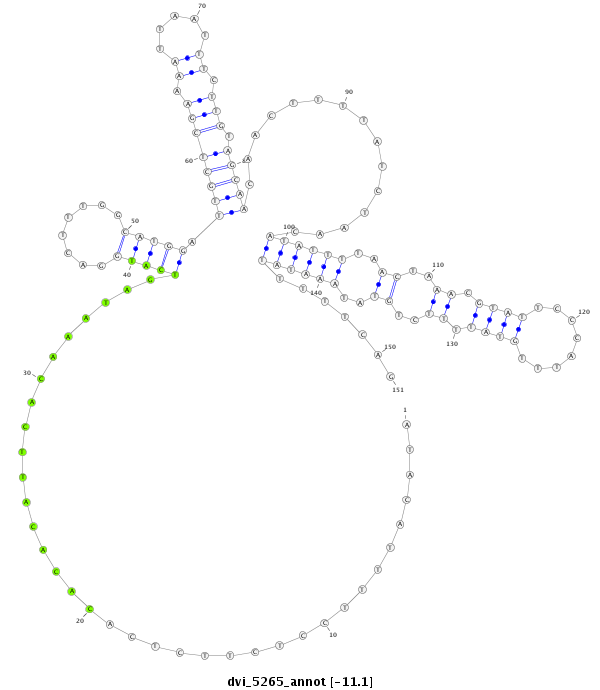 |
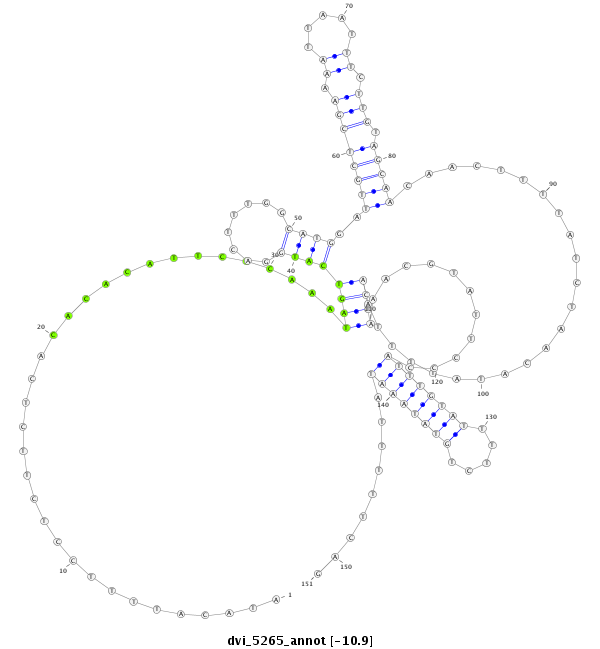 |
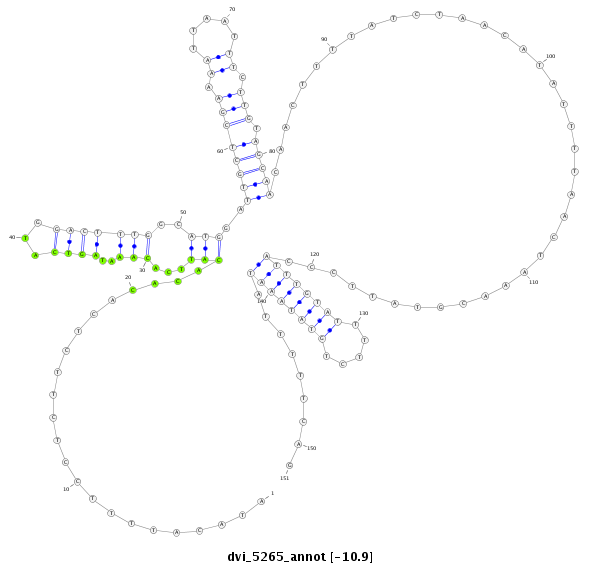 |
CDS [Dvir\GJ21446-cds]; exon [dvir_GLEANR_6804:3]; intron [Dvir\GJ21446-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------###########--------------------------------------- AAATTTTTAAAAGCTTGTATGCATAGAATACACAGATGAATACTGTTTGGATACATTTTCCTCTTCTCACACACATTCACAAATAGTCATGGACTTTGGCATGGATTGCTCGAAAATTAATTTCTTGTAGCAACAACTTTTATCTAACATATTTTAACTAAACGTATTCCCATTTGTATTTTCTGTATAAATATTTTTCAGGCATGATCTAATACCGGTTTGAGATAGTTCTGTTCGTTTATTGGCATTCC **************************************************....................................(((((........))))).((((((((.((.....)).))).)))))......................................((((((((.....)))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
GSM1528803 follicle cells |
SRR060666 160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................CACACATTCACAAATAGTCAT................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTAACGTATTCCCAT.............................................................................. | 15 | 1 | 18 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................ACAGGTGAATACTGGTGGG......................................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................GAAAAATAATCTCTTGTAGT........................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................CGAACAGATTTTAACTAA.......................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
TTTAAAAATTTTCGAACATACGTATCTTATGTGTCTACTTATGACAAACCTATGTAAAAGGAGAAGAGTGTGTGTAAGTGTTTATCAGTACCTGAAACCGTACCTAACGAGCTTTTAATTAAAGAACATCGTTGTTGAAAATAGATTGTATAAAATTGATTTGCATAAGGGTAAACATAAAAGACATATTTATAAAAAGTCCGTACTAGATTATGGCCAAACTCTATCAAGACAAGCAAATAACCGTAAGG
**************************************************....................................(((((........))))).((((((((.((.....)).))).)))))......................................((((((((.....)))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060677 Argx9_ovaries_total |
M047 female body |
SRR060686 Argx9_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
V116 male body |
SRR060661 160x9_0-2h_embryos_total |
SRR060664 9_males_carcasses_total |
SRR060658 140_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .TTAAAAATTTTCGAACATACGTATCT................................................................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................AAAATTGATTTGCATAAGGGTAAACAT......................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....AAAATTTTCGAAGATAAGTAT.................................................................................................................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CGAGCTGGTAATTAAAGAAC............................................................................................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................TAAAAGGAGGAGACTTTGTGTA............................................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CCAAACTTCATGAAGACAAG............... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................AAAAAGCCCGTACTATATT....................................... | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TATGTAAAAGGGGATGGGTGT.................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CAAGACAAGCAAATTCCTGT.... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................TTAAAAGGAGCAGAGTGT.................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:4738229-4738479 + | dvi_5265 | AAATTTTTAAAAGCTTGTATGCATAGAATACACAGATGAATACTGTTTGGATACATTTTCCTCTTCTCACACACATTCACAAATAGTCATGGACTTTGGCATGGATTGCTCGAAAATTAATTTCTTGTAGCAACAACTTTTATCTAACATATTTTAACTAAACGTATTCCCATTTGTATTTTCTGTATAAATATTTTTCAGGCATGATCTAATACCGGTTTGAGATAGTTCTGTTCGTTTATTGGCATTCC |
| droMoj3 | scaffold_6496:25329431-25329500 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCGCATTTATTTCTTTCAGGCATGATTTAACTTATCTTTGTGACAGTTCTGTTCGTCTTTTGATATCCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/16/2015 at 09:30 PM