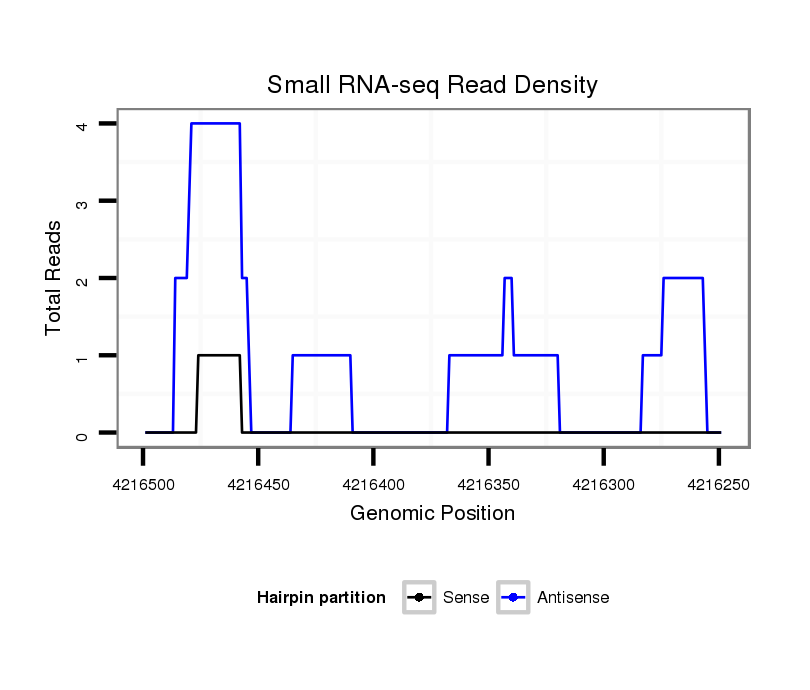
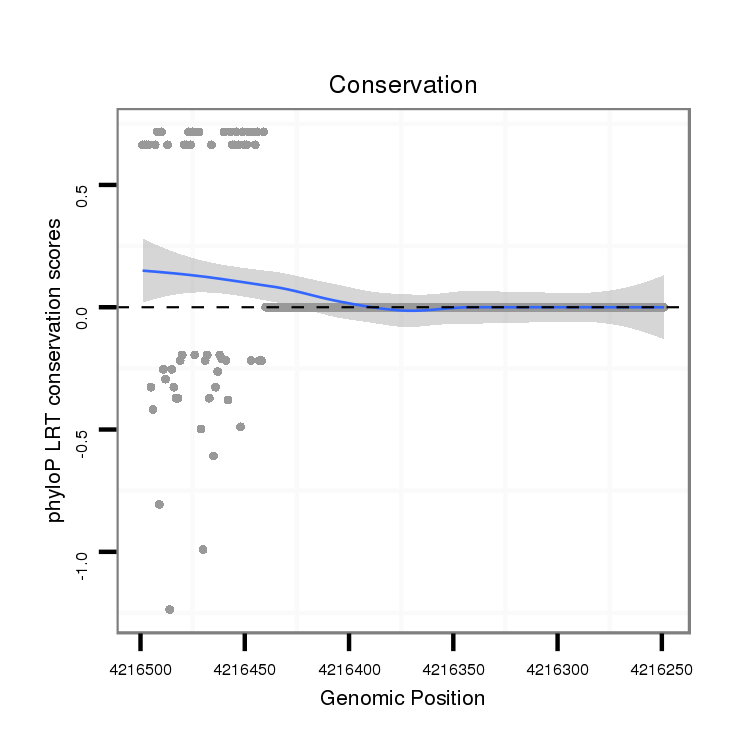
ID:dvi_5166 |
Coordinate:scaffold_12875:4216299-4216449 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_6324:3]; CDS [Dvir\GJ20920-cds]; intron [Dvir\GJ20920-in]; Antisense to intron [Dvir\GJ21415-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCGCACACAACGGACGGCGGCTGTCAATTGCGGGCAGTTCAAGGAGGTGGTGAGTAGACAGCTAGAGTTGGGTAAAATAGGGCAATGCACAATTCCTATCAGCTATATAATATACTAGTAGGAAGGTTATTTCATTAAAAATGATTTTTGATTTCGGTTACATTCAGACGAGAAAAAAAAACCCGAAATCATCCTAAAATGTAGAAATTCAGAAAAGCGTCTAAGGATCGAGCTGTATTTTGAAACGATT **************************************************.....((..((((....))))..))...(((((..........(((((((.((.((((...)))))).)))))))((((((((......))))))))..(((((((((.....(((......)))........))))))))))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
M047 female body |
V116 male body |
SRR060668 160x9_males_carcasses_total |
SRR060674 9x140_ovaries_total |
V053 head |
V047 embryo |
SRR060665 9_females_carcasses_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
SRR060667 160_females_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
SRR060684 140x9_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................GTCAATTGCGGGCAGTTCA................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AATTCAGGGAGGTAGTGA..................................................................................................................................................................................................... | 18 | 3 | 20 | 0.55 | 11 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AATTCAGGGAGGTGGTGAC.................................................................................................................................................................................................... | 19 | 3 | 7 | 0.43 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....ATAGAACGGACGGCGGCTGTT................................................................................................................................................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GTCCGACGATCGAGCTGTAT............ | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GTAGAAATTCAGACAACCGC.............................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AATTCAGGGAGGTAGTGAGT................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................AATTCAAGGAGGTAGTGA..................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AATTCCTATCAGCTTCATATT........................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TTCAAGGAGGTAGTGACA................................................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ....................................AGTTCAGGGAGGTAGTGA..................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GTCGGGCTTTATTTTGAAAC.... | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TGCGTCAAAGGATCGAGC................. | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................TGAAGAAATTCAAAAAAGCGC.............................. | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................CGCCTAGGGATCGAGTTGT.............. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TTCAAGGAGGTAGTGACAA.................................................................................................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................CAATTCAGGGAGGTAGTGA..................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TAGTTCAGGGAGGTAGTGA..................................................................................................................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ATTCAGGGAGGTGGTGAC.................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CCGCGTGTGTTGCCTGCCGCCGACAGTTAACGCCCGTCAAGTTCCTCCACCACTCATCTGTCGATCTCAACCCATTTTATCCCGTTACGTGTTAAGGATAGTCGATATATTATATGATCATCCTTCCAATAAAGTAATTTTTACTAAAAACTAAAGCCAATGTAAGTCTGCTCTTTTTTTTTGGGCTTTAGTAGGATTTTACATCTTTAAGTCTTTTCGCAGATTCCTAGCTCGACATAAAACTTTGCTAA
**************************************************.....((..((((....))))..))...(((((..........(((((((.((.((((...)))))).)))))))((((((((......))))))))..(((((((((.....(((......)))........))))))))))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM1528803 follicle cells |
M047 female body |
SRR060654 160x9_ovaries_total |
V053 head |
SRR060675 140x9_ovaries_total |
SRR060677 Argx9_ovaries_total |
SRR060681 Argx9_testes_total |
SRR060685 9xArg_0-2h_embryos_total |
V047 embryo |
SRR060660 Argentina_ovaries_total |
SRR060665 9_females_carcasses_total |
SRR060671 9x160_males_carcasses_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............CTGCCGCCGACAGTTAACGCCCGTCAAGT................................................................................................................................................................................................................. | 29 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................CCGACAGTTAACGCCCGTCAAGTTCC.............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................ATTATATGATCGTCCTTCCAATAAAGT.................................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................CCAATGTAAGTCTGCTCTTTTTTT....................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CCTAGCTCGACATAAAAC........ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................CGTGTTAAGGATAGTCGGTAT............................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................CGACAGTTAACGCCCGTCAAGTTCCT............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTAGTAGGAATTTACATCTTTAAGTCTT.................................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TCGCAGATTCCTAGCTCGACATAAAACT....... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TCTCAACCCATTTTATCCCGTTACGT................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CTGCCGCCGACCGTTGACGACCGTCAAGT................................................................................................................................................................................................................. | 29 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AGTAATTTTTACTAAAAACTAAAGCCAA........................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................TTTTTCTTGGGGTTTAGTAG......................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................CCGTTGAGTTCCTCCAACACT..................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................CCCAGTTTATGCCGTTTCGTGT............................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................TTATCCCGTTCCTTGTTAAGT.......................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GATCTAGTTCCTCCACCACT..................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTAAGGATAGTCGAAGTATAA........................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................CCTTTTTTTTTGGGCTTGGGT........................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................TCTGTCAATCTGAACCCAA................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:4216249-4216499 - | dvi_5166 | GGCGCACACAACGGACGGCGGCTGTCAATTGCGGGCAGTTCAAGGAGGTGGTGAGTAGACAGCTAGAGTTGGGTAAAATAGGGCAATGCACAATTCCTATCAGCTATATAATATACTAGTAGGAAGGTTATTTCATTAAAAATGATTTTTGATTTCGGTTACATTCAGACGAGAAAAAAAAACCCGAAATCATCCTAAAATGTAGAAATTCAGAAAAGCGTCTAAGGATCGAGCTGTATTTTGAAACGATT |
| droMoj3 | scaffold_6496:20836410-20836468 + | GGCGATCAGAGCGCGAAATGGCTGTCAAGAACAGCATGTTTAAGGAGGTGGTAAGTAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droGri2 | scaffold_15112:2025184-2025242 + | GGCGCTCAGAAAGAACAACAGCTGTTAATGGTAGGCAACTCCAGGAGCTGGTGAGTTGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/16/2015 at 08:09 PM