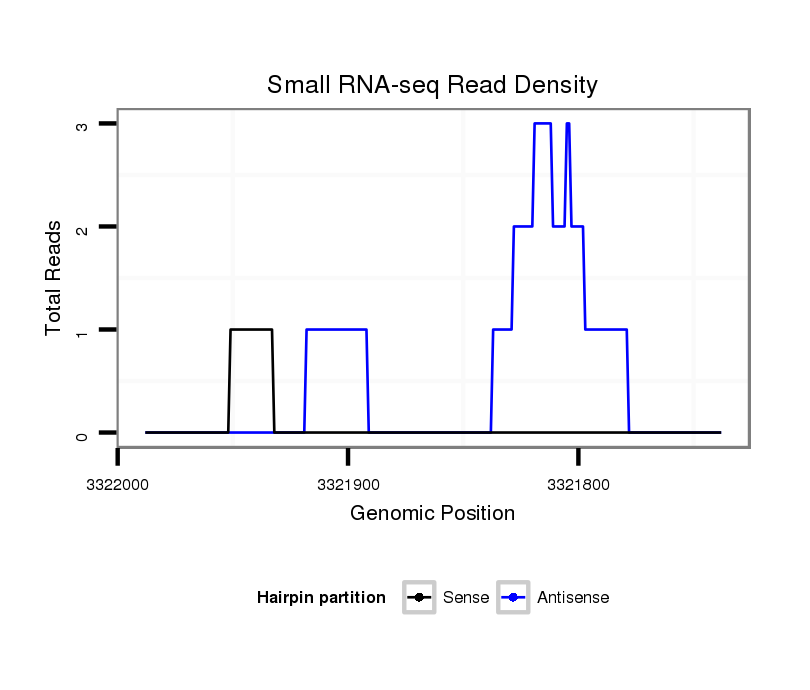
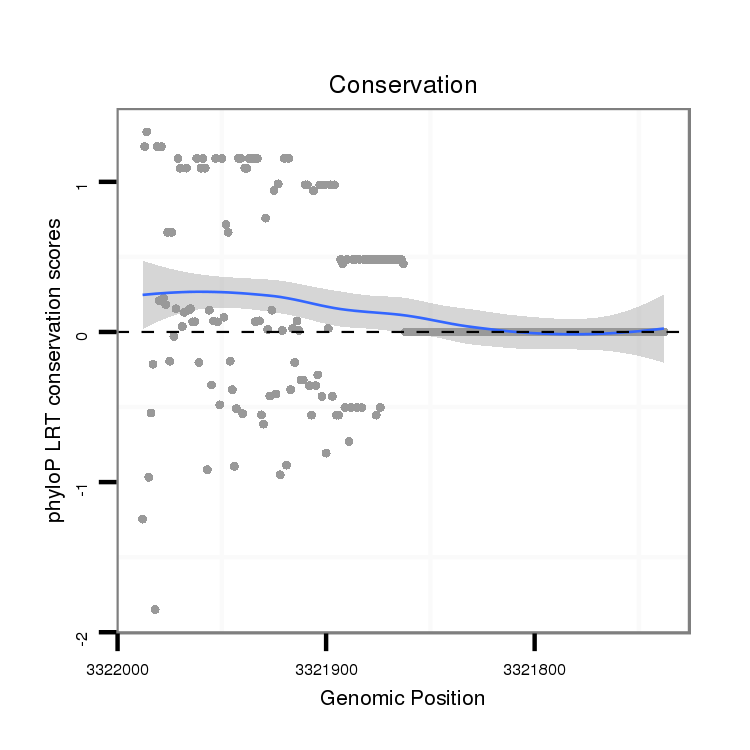
ID:dvi_5116 |
Coordinate:scaffold_12875:3321788-3321938 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dvir_GLEANR_6357:6]; CDS [Dvir\GJ20956-cds]; intron [Dvir\GJ20956-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGAAGCCGGCAGCGGAATGTCCTATATGCTGATCATTAACTGCTGTATCGGTAATTAAAACAACAAGCTCTATCTCAAATATCTATAAGAAAATATGTTACTAATTTTATTTTTCAAATTAATTTGCATAGATACATAGAGTAGTAGTTGTAGACTACGAAACACGCAAAGTCTAGCTTACCCTAAAACGCTTCCCATTGCGCGAATTTATTCTTCGGTATTTACTGTCCGATTAAACAAAATTTTCGGGT **************************************************..........((((..(((((((......(((((((..(((((((............)))))))((((....)))).))))))).)))))))....))))((((((.((.....))...))))))..............((........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060679 140x9_testes_total |
SRR060660 Argentina_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060672 9x160_females_carcasses_total |
SRR060668 160x9_males_carcasses_total |
SRR060669 160x9_females_carcasses_total |
SRR060687 9_0-2h_embryos_total |
SRR060676 9xArg_ovaries_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060659 Argentina_testes_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................GACTACGAAACCCACAGAGT............................................................................... | 20 | 3 | 5 | 1.20 | 6 | 1 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| .....................................AACTGCTGTATCGGTAATT................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AGCCGTTGTAGACTACGA........................................................................................... | 18 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ATTGCGCGAGTTTATTCCTCA.................................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................CTACGAAACCCACAGAGTC.............................................................................. | 19 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................TGACCCGAAATCGCTTCCC....................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................CTTCGCACTGCGCGAATTCA......................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................ACTACGAAACCCACAGAGT............................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| TGAAGAACGCAGCGGAATG........................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACTTCGGCCGTCGCCTTACAGGATATACGACTAGTAATTGACGACATAGCCATTAATTTTGTTGTTCGAGATAGAGTTTATAGATATTCTTTTATACAATGATTAAAATAAAAAGTTTAATTAAACGTATCTATGTATCTCATCATCAACATCTGATGCTTTGTGCGTTTCAGATCGAATGGGATTTTGCGAAGGGTAACGCGCTTAAATAAGAAGCCATAAATGACAGGCTAATTTGTTTTAAAAGCCCA
**************************************************..........((((..(((((((......(((((((..(((((((............)))))))((((....)))).))))))).)))))))....))))((((((.((.....))...))))))..............((........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060665 9_females_carcasses_total |
SRR060677 Argx9_ovaries_total |
SRR060678 9x140_testes_total |
SRR060676 9xArg_ovaries_total |
SRR060667 160_females_carcasses_total |
SRR060657 140_testes_total |
M028 head |
SRR060662 9x160_0-2h_embryos_total |
V116 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................TTGTTGTTCGAGATTAAGTTT............................................................................................................................................................................ | 21 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TCTGATGCTTTGTGCGTTTCAGATCG.......................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................TCAGATCGAATGGGATTTTGCG............................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................ATTTTGCGAAGGGTAACGCGCTTAAAT......................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................TTGTGCGTTTCAGATCGAATGGGAT.................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ATAGAGTTTATAGATATTCTTTTATAC.......................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GCGATGGCTAACGCGCTT............................................. | 18 | 2 | 5 | 0.60 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| ...............TTACCGGATATACGAGTTGTAA...................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TGTTGTTCGAGATTAAGTTTG........................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTAGTAGTAATTGACGAGAT............................................................................................................................................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................TTTTCGAGATAGAGTTGA........................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................AGTTTGTTCTGCGAGATAG................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:3321738-3321988 - | dvi_5116 | TGAAGCCGGCAGCGGAATGTCCTATA---TGCTGATCATTAACTGCTGTATCGGTAATTAAAACAACA---AGCTCTATCTCAAATAT-----CTATAAGAA--AATATGTTACTAATTTTATTTTTCAA-------ATTAATTTGCATAGATACATAGAGTAGTAGTTGTAGACTACGAAACACGCAAAGTCTAGCTTACCCTAAAACGCTTCCCATTGCGCGAATTTATTCTTCGGTATTTACTGTCCGATTAAACAAAATTTTCGGGT |
| droMoj3 | scaffold_6496:21821083-21821218 + | GGACCCGGGCCGCGGAATGGCCTACG---TGCTGCTTTTCTATTGCACTATTGGTAACTAAC-GGA------ATTTTTTCATGGATGACAATTCCGTTAAAAGTTAATTGCTCTTAGTCTTATTTATTAATTTATCTATTAATTTG----------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15112:2822856-2822932 + | GGAACATGACAGCAGCATGTCCTATA---TACTGTTTATTTACTGTTCCATTGGTAATTACA-CAACT---ATTTATACTTTAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454039:1444201-1444212 + | CGACTTGGGCAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4845:10307631-10307722 + | AGATTTCGGCAA---AGTGTTCAGTATTATACTGTACATTAAC---ATGATAGGTAATTCGGACTTCACTGAATTCTTTTTTGGATA------CTGTAACGA--AA--------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/16/2015 at 07:57 PM