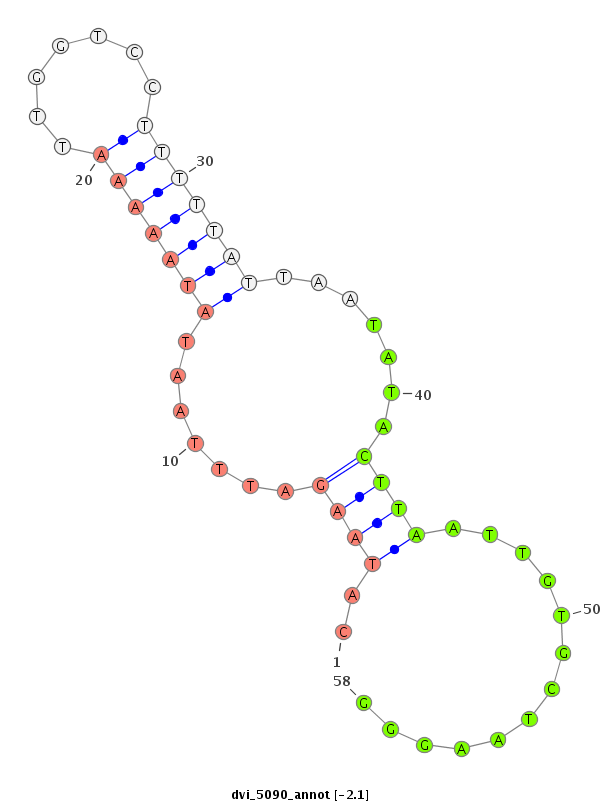
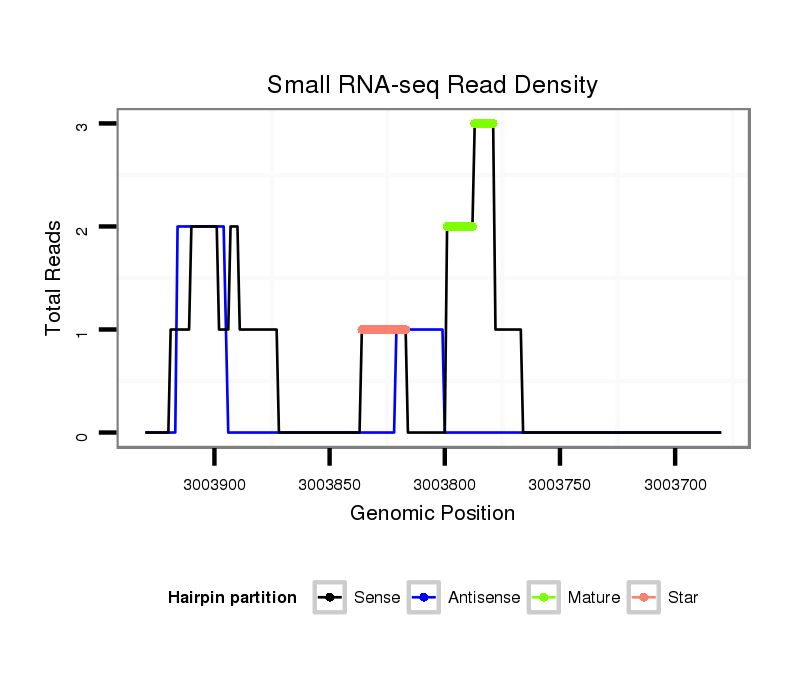
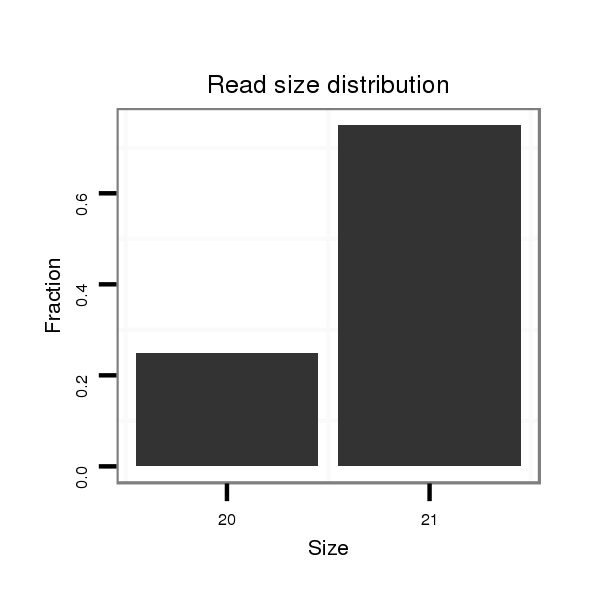
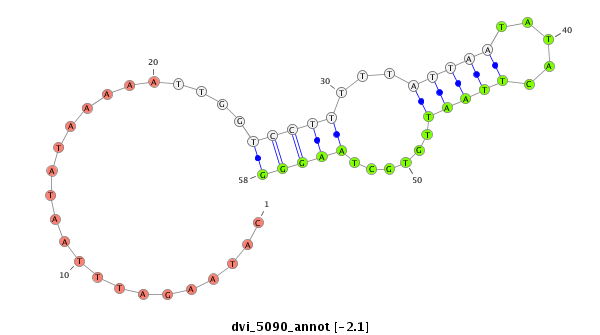
ID:dvi_5090 |
Coordinate:scaffold_12875:3003730-3003880 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
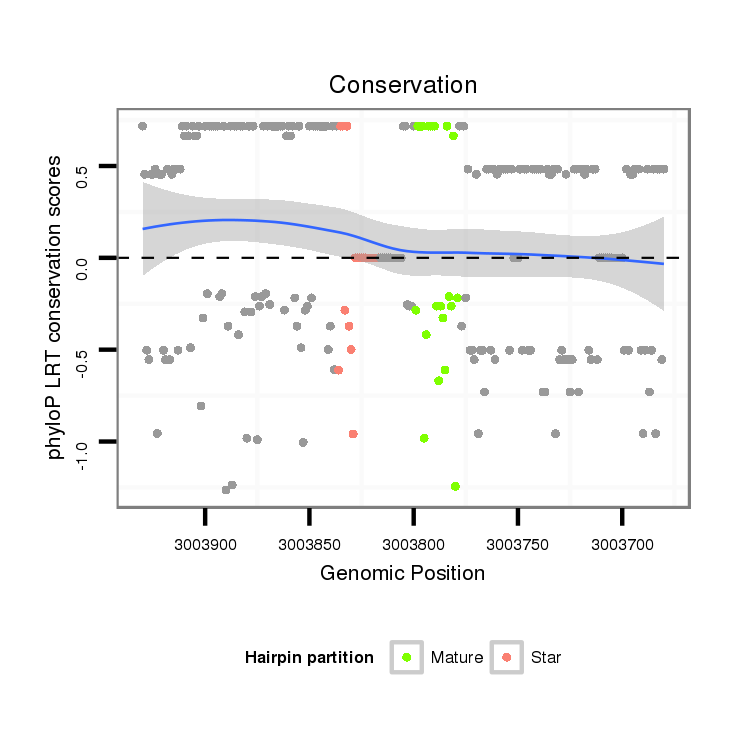
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -2.1 | -2.0 | -2.0 |
 |
 |
 |
exon [dvir_GLEANR_6378:1]; CDS [Dvir\GJ20978-cds]; intron [Dvir\GJ20978-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGCACGACGCAATTGAAGTTGTGCAGCTCCTGATAATACTGCTGATTTTCGTGATAATACTAAAAATAACTGATGTGTATTATTAAATAAGTGTCATAAGATTTAATATAAAAATTGGTCCTTTTTATTAATATACTTAATTGTGCTAAGGGAGTCTGATGGGCTTACAAGTAAATCTTAGAGATGCATTTAACTGCAGAACAGATAATTTACACTTATAAGTTGTAAATGTATCGATAGGCAGAAGTTTT **********************************************************************************************..((((.......(((((((.......))))))).......)))).............*************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V116 male body |
M047 female body |
SRR060678 9x140_testes_total |
SRR060655 9x160_testes_total |
SRR060657 140_testes_total |
SRR060659 Argentina_testes_total |
SRR060679 140x9_testes_total |
SRR060686 Argx9_0-2h_embryos_total |
SRR060687 9_0-2h_embryos_total |
SRR060662 9x160_0-2h_embryos_total |
SRR060666 160_males_carcasses_total |
M027 male body |
SRR060658 140_ovaries_total |
V053 head |
SRR060684 140x9_0-2h_embryos_total |
SRR060665 9_females_carcasses_total |
M028 head |
SRR060669 160x9_females_carcasses_total |
SRR060660 Argentina_ovaries_total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................GTCCTAAGGGAGGCTCATGG......................................................................................... | 20 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GTCCTAAGGGGGTCTCATGG......................................................................................... | 20 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TGTCCTAAGGGGGTCTCATGG......................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TATACTTAATTGTGCTAAGGG................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TGCTAAGGGAGTCTGATGGGC....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TGTCCTAAGGGGGTCTCATG.......................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TGTGCTAAGGGGGGCTCATGG......................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GTCCTAAGGGGGGCTGATGG......................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CATAAGATTTAATATAAAAA......................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........ATTGAAGTTGTGCAGCTCCTG........................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ACTGCTGATTTTCGTGATAAT................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GTGCAGCTCCTGATAATACTG.................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CTAAGGGGGGCTCATGGG........................................................................................ | 18 | 3 | 18 | 0.72 | 13 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 6 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| ..............................................................................................................................................GTCCTAAGGGGGTCTCATG.......................................................................................... | 19 | 3 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TAATTGTGCTAGGGGTGTCTT............................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CGGCTCCTGATAGTACTGC................................................................................................................................................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TAAGGGGGGCTGATGGGC....................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GTCCTAAGGGAGGCTCATG.......................................................................................... | 19 | 3 | 7 | 0.29 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TGCTAAGGGAGTATGCGG.......................................................................................... | 18 | 3 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................TCAGGGATGCGTTTAACTGC...................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....GACGCTATTGAAATTGTGAAG................................................................................................................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GTCCTAAGGGGGTCTCAT........................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................ACTGCTGATTTTCGCGTTCA.................................................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ATATAATTAATTGTGGTAAGA.................................................................................................... | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ACGTGCTGCGTTAACTTCAACACGTCGAGGACTATTATGACGACTAAAAGCACTATTATGATTTTTATTGACTACACATAATAATTTATTCACAGTATTCTAAATTATATTTTTAACCAGGAAAAATAATTATATGAATTAACACGATTCCCTCAGACTACCCGAATGTTCATTTAGAATCTCTACGTAAATTGACGTCTTGTCTATTAAATGTGAATATTCAACATTTACATAGCTATCCGTCTTCAAAA
***************************************************************************************************..((((.......(((((((.......))))))).......)))).............********************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR060678 9x140_testes_total |
SRR060680 9xArg_testes_total |
SRR060688 160_ovaries_total |
SRR060671 9x160_males_carcasses_total |
SRR060682 9x140_0-2h_embryos_total |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............CTTCAACACGTCGAGGACTAT........................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............CCTTCAACACGTCGAGGACTAT........................................................................................................................................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............CTTCAACACGTCGAGGACTATT....................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................TTTTTAACCAGGAAAAATAAT......................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................CTGGTATGACGACTACAAGC........................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................ACACGTGGAGGAATATTA...................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droVir3 | scaffold_12875:3003680-3003930 - | dvi_5090 | TGCACGACGCAATTGAAGTTGTGCAGCTCCTGATAATACTGCTGATTTTCGTGATAATACTAAAAATAACTGATGTGTATTATTAAATAA------GTGTCAT--------------------------------------------------------------------------AA-----------------------------GATTTAATATAAAAATTGGTCCTTTTTATTAATATACTTA-----------ATTGTGCTAAGGGAGTC-------------TGATGG----------------------GCTTACAAGTAAATCTTAGAGATGCATTTAACTGCAGAACAGATAATTTACACTTATAAGTTGTAAATGTATCGATAGGCAGAAGTTTT |
| droMoj3 | scaffold_6496:22190087-22190448 + | TGTTCGAGGCGTTAGAAATTGTGCAGCTGATGATAATACTCTTCATATTCATGATTTTACTGAAAATAGCTGATATGGGATATTAAATAC------ATCTGATTGTCAACTTATGGGCATTTGTTTAATATATGTACATAGGAAATGCTGAGGTGCTAATTGGGCAAAAGATTTCATGATTTTTGGTAGTCGTAAAATCTGCAATCGTACA-----------------------TACATACATATATAGAGATGTGTCTATATATGTATGTAAATTTTTTAGTCGACTCTAGAGCCACGTGTAACATGAATGTATGTATGTATATGTAAATTT---AAATATATTTACATGCACATTTGTGTATGTACATATAA------------CACCGATAACTATGACTTAT | |
| droGri2 | scaffold_15112:3152659-3152790 + | T------------------TGTGGAGCTGCTAATAATGTTTTTAATATTATTTACCACATTAAAAATAGCTGAAATCAGTTTTTAAATAATAAAAAATGTCAT--------------------------------------------------------------------------GA-----------------------------AAC-----------------------TATTAACATAAATAGCCTTGCATATATTTTGCTGAGCGAATC---------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 01:17 PM